Effects of pH on proteins: predictions for ensemble and single-molecule pulling experiments
- PMID: 22148729
- PMCID: PMC3262061
- DOI: 10.1021/ja206557y
Effects of pH on proteins: predictions for ensemble and single-molecule pulling experiments
Abstract
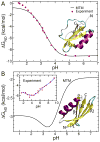
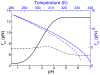
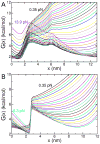
Protein conformations change among distinct thermodynamic states as solution conditions (temperature, denaturants, pH) are altered or when they are subjected to mechanical forces. A quantitative description of the changes in the relative stabilities of the various thermodynamic states is needed to interpret and predict experimental outcomes. We provide a framework based on the Molecular Transfer Model (MTM) to account for pH effects on the properties of globular proteins. The MTM utilizes the partition function of a protein calculated from molecular simulations at one set of solution conditions to predict protein properties at another set of solution conditions. To take pH effects into account, we utilized experimentally measured pK(a) values in the native and unfolded states to calculate the free energy of transferring a protein from a reference pH to the pH of interest. We validate our approach by demonstrating that the native-state stability as a function of pH is accurately predicted for chymotrypsin inhibitor 2 (CI2) and protein G. We use the MTM to predict the response of CI2 and protein G subjected to a constant force (f) and varying pH. The phase diagrams of CI2 and protein G as a function of f and pH are dramatically different and reflect the underlying pH-dependent stability changes in the absence of force. The calculated equilibrium free energy profiles as functions of the end-to-end distance of the two proteins show that, at various pH values, CI2 unfolds via an intermediate when subjected to f. The locations of the two transition states move toward the more unstable state as f is changed, which is in accord with the Hammond-Leffler postulate. In sharp contrast, force-induced unfolding of protein G occurs in a single step. Remarkably, the location of the transition state with respect to the folded state is independent of f, which suggests that protein G is mechanically brittle. The MTM provides a natural framework for predicting the outcomes of ensemble and single-molecule experiments for a wide range of solution conditions.
© 2011 American Chemical Society
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

