Uptake of radiolabeled GlcNAc into Saccharomyces cerevisiae via native hexose transporters and its in vivo incorporation into GPI precursors in cells expressing heterologous GlcNAc kinase
- PMID: 22151002
- PMCID: PMC3498731
- DOI: 10.1111/j.1567-1364.2011.00778.x
Uptake of radiolabeled GlcNAc into Saccharomyces cerevisiae via native hexose transporters and its in vivo incorporation into GPI precursors in cells expressing heterologous GlcNAc kinase
Abstract
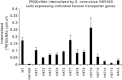
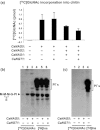
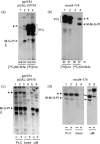
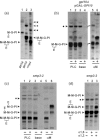
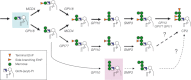
Yeast glycan biosynthetic pathways are commonly studied through metabolic incorporation of an exogenous radiolabeled compound into a target glycan. In Saccharomyces cerevisiae glycosylphosphatidylinositol (GPI) biosynthesis, [(3) H]inositol has been widely used to identify intermediates that accumulate in conditional GPI synthesis mutants. However, this approach also labels non-GPI lipid species that overwhelm detection of early GPI intermediates during chromatography. In this study, we show that despite lacking the ability to metabolize N-acetylglucosamine (GlcNAc), S. cerevisiae is capable of importing low levels of extracellular GlcNAc via almost all members of the hexose transporter family. Furthermore, expression of a heterologous GlcNAc kinase gene permits efficient incorporation of exogenous [(14) C]GlcNAc into nascent GPI structures in vivo, dramatically lowering the background signal from non-GPI lipids. Utilizing this new method with several conditional GPI biosynthesis mutants, we observed and characterized novel accumulating lipids that were not previously visible using [(3) H]inositol labeling. Chemical and enzymatic treatments of these lipids indicated that each is a GPI intermediate likely having one to three mannoses and lacking ethanolamine phosphate (Etn-P) side-branches. Our data support a model of yeast GPI synthesis that bifurcates after the addition of the first mannose and that includes a novel branch that produces GPI species lacking Etn-P side-branches.
© 2011 Federation of European Microbiological Societies. Published by Blackwell Publishing Ltd. All rights reserved.
Figures

References
-
- Benachour A, Sipos G, Flury I, et al. Deletion of GPI7, a yeast gene required for addition of a side chain to the glycosylphosphatidylinositol (GPI) core structure, affects GPI protein transport, remodeling, and cell wall integrity. J Biol Chem. 1999;274:15251–15261. - PubMed
-
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

