Human papillomavirus L2 facilitates viral escape from late endosomes via sorting nexin 17
- PMID: 22151726
- PMCID: PMC3276720
- DOI: 10.1111/j.1600-0854.2011.01320.x
Human papillomavirus L2 facilitates viral escape from late endosomes via sorting nexin 17
Abstract
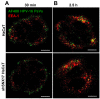
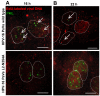
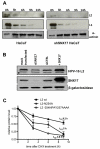
The human papillomavirus (HPV) L2 capsid protein plays an essential role during the early stages of viral infection, but the molecular mechanisms underlying its mode of action remain obscure. Using a proteomic approach, we have identified the adaptor protein, sorting nexin 17 (SNX17) as a strong interacting partner of HPV L2. This interaction occurs through a highly conserved SNX17 consensus binding motif, which is present in the majority of HPV L2 proteins analysed. Using mutants of L2 defective for SNX17 interaction, or siRNA ablation of SNX17 expression, we demonstrate that the interaction between L2 and SNX17 is essential for viral infection. Furthermore, loss of the L2-SNX17 interaction results in enhanced turnover of the L2 protein and decreased stability of the viral capsids, and concomitantly, there is a dramatic decrease in the efficiency with which viral genomes transit to the nucleus. Indeed, using a range of endosomal and lysosomal markers, we show that capsids defective in their capacity to bind SNX17 transit much more rapidly to the lysosomal compartment. These results demonstrate that the L2-SNX17 interaction is essential for viral infection and facilitates the escape of the L2-DNA complex from the late endosomal/lysosomal compartments.
© 2011 John Wiley & Sons A/S.
Figures

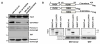


References
-
- Smith JS, Lindsay L, Hoots B, Keys J, Franceschi S, Winer R, Clifford GM. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: a meta-analysis update. Int J Cancer. 2007;121:621–632. - PubMed
-
- Pereira R, Hitzeroth II, Rybicki EP. Insights into the role and function of L2, the minor capsid protein of papillomaviruses. Arch Virol. 2009;154:187–197. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

