The prototype HIV-1 maturation inhibitor, bevirimat, binds to the CA-SP1 cleavage site in immature Gag particles
- PMID: 22151792
- PMCID: PMC3267693
- DOI: 10.1186/1742-4690-8-101
The prototype HIV-1 maturation inhibitor, bevirimat, binds to the CA-SP1 cleavage site in immature Gag particles
Abstract
Background: Bevirimat, the prototype Human Immunodeficiency Virus type 1 (HIV-1) maturation inhibitor, is highly potent in cell culture and efficacious in HIV-1 infected patients. In contrast to inhibitors that target the active site of the viral protease, bevirimat specifically inhibits a single cleavage event, the final processing step for the Gag precursor where p25 (CA-SP1) is cleaved to p24 (CA) and SP1.
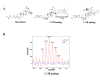
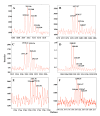
Results: In this study, photoaffinity analogs of bevirimat and mass spectrometry were employed to map the binding site of bevirimat to Gag within immature virus-like particles. Bevirimat analogs were found to crosslink to sequences overlapping, or proximal to, the CA-SP1 cleavage site, consistent with previous biochemical data on the effect of bevirimat on Gag processing and with genetic data from resistance mutations, in a region predicted by NMR and mutational studies to have α-helical character. Unexpectedly, a second region of interaction was found within the Major Homology Region (MHR). Extensive prior genetic evidence suggests that the MHR is critical for virus assembly.
Conclusions: This is the first demonstration of a direct interaction between the maturation inhibitor, bevirimat, and its target, Gag. Information gained from this study sheds light on the mechanisms by which the virus develops resistance to this class of drug and may aid in the design of next-generation maturation inhibitors.
Figures




Similar articles
-
Resistance to Second-Generation HIV-1 Maturation Inhibitors.J Virol. 2019 Mar 5;93(6):e02017-18. doi: 10.1128/JVI.02017-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30567982 Free PMC article.
-
HIV-1 protease inhibitor mutations affect the development of HIV-1 resistance to the maturation inhibitor bevirimat.Retrovirology. 2011 Aug 24;8:70. doi: 10.1186/1742-4690-8-70. Retrovirology. 2011. PMID: 21864346 Free PMC article.
-
Alkyl Amine Bevirimat Derivatives Are Potent and Broadly Active HIV-1 Maturation Inhibitors.Antimicrob Agents Chemother. 2015 Oct 19;60(1):190-7. doi: 10.1128/AAC.02121-15. Print 2016 Jan. Antimicrob Agents Chemother. 2015. PMID: 26482309 Free PMC article.
-
Bevirimat: a novel maturation inhibitor for the treatment of HIV-1 infection.Antivir Chem Chemother. 2008;19(3):107-13. doi: 10.1177/095632020801900301. Antivir Chem Chemother. 2008. PMID: 19024627 Review.
-
HIV-1 Maturation: Lessons Learned from Inhibitors.Viruses. 2020 Aug 26;12(9):940. doi: 10.3390/v12090940. Viruses. 2020. PMID: 32858867 Free PMC article. Review.
Cited by
-
Structural and functional insights into the HIV-1 maturation inhibitor binding pocket.PLoS Pathog. 2012;8(11):e1002997. doi: 10.1371/journal.ppat.1002997. Epub 2012 Nov 8. PLoS Pathog. 2012. PMID: 23144615 Free PMC article.
-
Fullerene Derivatives Strongly Inhibit HIV-1 Replication by Affecting Virus Maturation without Impairing Protease Activity.Antimicrob Agents Chemother. 2016 Sep 23;60(10):5731-41. doi: 10.1128/AAC.00341-16. Print 2016 Oct. Antimicrob Agents Chemother. 2016. PMID: 27431232 Free PMC article.
-
Review of Current Cell-Penetrating Antibody Developments for HIV-1 Therapy.Molecules. 2018 Feb 6;23(2):335. doi: 10.3390/molecules23020335. Molecules. 2018. PMID: 29415435 Free PMC article. Review.
-
Structural biology of supramolecular assemblies by magic-angle spinning NMR spectroscopy.Q Rev Biophys. 2017 Jan;50:e1. doi: 10.1017/S0033583516000159. Q Rev Biophys. 2017. PMID: 28093096 Free PMC article. Review.
-
MAS NMR of HIV-1 protein assemblies.J Magn Reson. 2015 Apr;253:10-22. doi: 10.1016/j.jmr.2014.12.009. J Magn Reson. 2015. PMID: 25797001 Free PMC article.
References
-
- Gallant JE. Antiretroviral drug resistance and resistance testing. Top HIV Med. 2005;13:138–142. - PubMed
-
- Li F, Goila-Gaur R, Salzwedel K, Kilgore NR, Reddick M, Matallana C, Castillo A, Zoumplis D, Martin DE, Orenstein JM, Allaway GP, Freed EO, Wild CT. PA-457: A potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc Nat'l Acad Sci USA. 2003;100:13555–13560. doi: 10.1073/pnas.2234683100. - DOI - PMC - PubMed
-
- Kanamoto T, Kashiwada Y, Kanbara K, Gotoh K, Yoshimori M, Goto T, Sano K, Nakashima H. Anti-human immunodeficiency virus activity of YK-FH312 (a betulinic acid derivative), a novel compound blocking viral maturation. Antimicrob Agents Chemother. 2001;45:1225–1230. doi: 10.1128/AAC.45.4.1225-1230.2001. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

