De novo assembly of Euphorbia fischeriana root transcriptome identifies prostratin pathway related genes
- PMID: 22151917
- PMCID: PMC3273484
- DOI: 10.1186/1471-2164-12-600
De novo assembly of Euphorbia fischeriana root transcriptome identifies prostratin pathway related genes
Abstract
Background: Euphorbia fischeriana is an important medicinal plant found in Northeast China. The plant roots contain many medicinal compounds including 12-deoxyphorbol-13-acetate, commonly known as prostratin that is a phorbol ester from the tigliane diterpene series. Prostratin is a protein kinase C activator and is effective in the treatment of Human Immunodeficiency Virus (HIV) by acting as a latent HIV activator. Latent HIV is currently the biggest limitation for viral eradication. The aim of this study was to sequence, assemble and annotate the E. fischeriana transcriptome to better understand the potential biochemical pathways leading to the synthesis of prostratin and other related diterpene compounds.
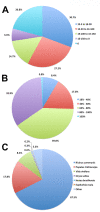
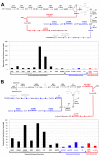
Results: In this study we conducted a high throughput RNA-seq approach to sequence the root transcriptome of E. fischeriana. We assembled 18,180 transcripts, of these the majority encoded protein-coding genes and only 17 transcripts corresponded to known RNA genes. Interestingly, we identified 5,956 protein-coding transcripts with high similarity (> = 75%) to Ricinus communis, a close relative to E. fischeriana. We also evaluated the conservation of E. fischeriana genes against EST datasets from the Euphorbeacea family, which included R. communis, Hevea brasiliensis and Euphorbia esula. We identified a core set of 1,145 gene clusters conserved in all four species and 1,487 E. fischeriana paralogous genes. Furthermore, we screened E. fischeriana transcripts against an in-house reference database for genes implicated in the biosynthesis of upstream precursors to prostratin. This identified 24 and 9 candidate transcripts involved in the terpenoid and diterpenoid biosyntehsis pathways, respectively. The majority of the candidate genes in these pathways presented relatively low expression levels except for 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase (HDS) and isopentenyl diphosphate/dimethylallyl diphosphate synthase (IDS), which are required for multiple downstream pathways including synthesis of casbene, a proposed precursor to prostratin.
Conclusion: The resources generated in this study provide new insights into the upstream pathways to the synthesis of prostratin and will likely facilitate functional studies aiming to produce larger quantities of this compound for HIV research and/or treatment of patients.
Figures
Similar articles
-
Esterification with a Long-Chain Fatty Acid Elevates the Exposure Toxicity of Tigliane Diterpenoids from Euphorbia fischeriana Roots against Nematodes.J Agric Food Chem. 2023 Aug 30;71(34):12730-12740. doi: 10.1021/acs.jafc.3c03460. Epub 2023 Aug 21. J Agric Food Chem. 2023. PMID: 37599642
-
[Euphorbia fischeriana extract reactivates latent HIV through nuclear factor-κB pathway].Nan Fang Yi Ke Da Xue Xue Bao. 2015 Nov;35(11):1614-8. Nan Fang Yi Ke Da Xue Xue Bao. 2015. PMID: 26607086 Chinese.
-
A new highly oxygenated abietane diterpenoid and a new lysosome generating phorbol ester from the roots of Euphorbia fischeriana Steud.Nat Prod Res. 2020 Nov;34(21):3027-3035. doi: 10.1080/14786419.2019.1607331. Epub 2019 May 14. Nat Prod Res. 2020. PMID: 31084207
-
Prostratin: An Overview.Mini Rev Med Chem. 2015;15(13):1122-30. doi: 10.2174/1389557515666150511154108. Mini Rev Med Chem. 2015. PMID: 25963564 Review.
-
Existing knowledge on Euphorbia fischeriana Steud. (Euphorbiaceae): Traditional uses, clinical applications, phytochemistry, pharmacology and toxicology.J Ethnopharmacol. 2021 Jul 15;275:114095. doi: 10.1016/j.jep.2021.114095. Epub 2021 Apr 2. J Ethnopharmacol. 2021. PMID: 33819505
Cited by
-
Deep sequencing reveals transcriptome re-programming of Taxus × media cells to the elicitation with methyl jasmonate.PLoS One. 2013 Apr 30;8(4):e62865. doi: 10.1371/journal.pone.0062865. Print 2013. PLoS One. 2013. PMID: 23646152 Free PMC article.
-
High-throughput sequencing analysis of Euphorbia fischeriana Steud provides insights into the molecular mechanism of pharmaceutical ingredient biosynthesis.3 Biotech. 2018 Oct;8(10):449. doi: 10.1007/s13205-018-1475-9. Epub 2018 Oct 12. 3 Biotech. 2018. PMID: 30333951 Free PMC article.
-
Transcriptome and Metabolome Analysis of the Synthesis Pathways of Allelochemicals in Eupatorium adenophorum.ACS Omega. 2022 May 4;7(19):16803-16816. doi: 10.1021/acsomega.2c01816. eCollection 2022 May 17. ACS Omega. 2022. PMID: 35601343 Free PMC article.
-
Characterization of Withania somnifera leaf transcriptome and expression analysis of pathogenesis-related genes during salicylic acid signaling.PLoS One. 2014 Apr 16;9(4):e94803. doi: 10.1371/journal.pone.0094803. eCollection 2014. PLoS One. 2014. PMID: 24739900 Free PMC article.
-
Comparative transcriptome analysis of Gastrodia elata (Orchidaceae) in response to fungus symbiosis to identify gastrodin biosynthesis-related genes.BMC Genomics. 2016 Mar 9;17:212. doi: 10.1186/s12864-016-2508-6. BMC Genomics. 2016. PMID: 26960548 Free PMC article.
References
-
- Cox PA. Ensuring equitable benefits: The Falealupo covenant and the isolation of anti-viral drug prostratin from a Samoan medicinal plant. Pharm Biol. 2001;39:33–40. - PubMed
-
- Gustafson KR, Cardellina JH, McMahon JB, Gulakowski RJ, Ishitoya J, Szallasi Z, Lewin NE, Blumberg PM, Weislow OS, Beutler JA, Buckheit RW, Cragg GM, Cox PA, Bader JP, Boyd MR. A nonpromoting phorbol from the samoan medicinal plant Homalanthus nutans inhibits cell killing by HIV-1. J Med Chem. 1992;35(11):1978–1986. doi: 10.1021/jm00089a006. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

