Cloning and characterization of a pectin lyase gene from Colletotrichum lindemuthianum and comparative phylogenetic/structural analyses with genes from phytopathogenic and saprophytic/opportunistic microorganisms
- PMID: 22151976
- PMCID: PMC3271051
- DOI: 10.1186/1471-2180-11-260
Cloning and characterization of a pectin lyase gene from Colletotrichum lindemuthianum and comparative phylogenetic/structural analyses with genes from phytopathogenic and saprophytic/opportunistic microorganisms
Abstract
Background: Microorganisms produce cell-wall-degrading enzymes as part of their strategies for plant invasion/nutrition. Among these, pectin lyases (PNLs) catalyze the depolymerization of esterified pectin by a β-elimination mechanism. PNLs are grouped together with pectate lyases (PL) in Family 1 of the polysaccharide lyases, as they share a conserved structure in a parallel β-helix. The best-characterized fungal pectin lyases are obtained from saprophytic/opportunistic fungi in the genera Aspergillus and Penicillium and from some pathogens such as Colletotrichum gloeosporioides.The organism used in the present study, Colletotrichum lindemuthianum, is a phytopathogenic fungus that can be subdivided into different physiological races with different capacities to infect its host, Phaseolus vulgaris. These include the non-pathogenic and pathogenic strains known as races 0 and 1472, respectively.
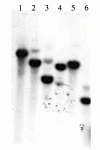
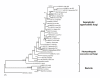
Results: Here we report the isolation and sequence analysis of the Clpnl2 gene, which encodes the pectin lyase 2 of C. lindemuthianum, and its expression in pathogenic and non-pathogenic races of C. lindemuthianum grown on different carbon sources. In addition, we performed a phylogenetic analysis of the deduced amino acid sequence of Clpnl2 based on reported sequences of PNLs from other sources and compared the three-dimensional structure of Clpnl2, as predicted by homology modeling, with those of other organisms. Both analyses revealed an early separation of bacterial pectin lyases from those found in fungi and oomycetes. Furthermore, two groups could be distinguished among the enzymes from fungi and oomycetes: one comprising enzymes from mostly saprophytic/opportunistic fungi and the other formed mainly by enzymes from pathogenic fungi and oomycetes. Clpnl2 was found in the latter group and was grouped together with the pectin lyase from C. gloeosporioides.
Conclusions: The Clpnl2 gene of C. lindemuthianum shares the characteristic elements of genes coding for pectin lyases. A time-course analysis revealed significant differences between the two fungal races in terms of the expression of Clpnl2 encoding for pectin lyase 2. According to the results, pectin lyases from bacteria and fungi separated early during evolution. Likewise, the enzymes from fungi and oomycetes diverged in accordance with their differing lifestyles. It is possible that the diversity and nature of the assimilatory carbon substrates processed by these organisms played a determinant role in this phenomenon.
Figures


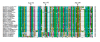


Similar articles
-
Evolution and functional characterization of pectate lyase PEL12, a member of a highly expanded Clonostachys rosea polysaccharide lyase 1 family.BMC Microbiol. 2018 Nov 7;18(1):178. doi: 10.1186/s12866-018-1310-9. BMC Microbiol. 2018. PMID: 30404596 Free PMC article.
-
The pectate lyase encoded by the pecCl1 gene is an important determinant for the aggressiveness of Colletotrichum lindemuthianum.J Microbiol. 2013 Aug;51(4):461-70. doi: 10.1007/s12275-013-3078-9. Epub 2013 Aug 30. J Microbiol. 2013. PMID: 23990297
-
A comparison of the pectate lyase genes, pel-1 and pel-2, of Colletotrichum gloeosporioides f.sp. malvae and the relationship between their expression in culture and during necrotrophic infection.Gene. 2000 Feb 8;243(1-2):139-50. doi: 10.1016/s0378-1119(99)00546-6. Gene. 2000. PMID: 10675622
-
Bacterial pectate lyases, structural and functional diversity.Environ Microbiol Rep. 2014 Oct;6(5):427-40. doi: 10.1111/1758-2229.12166. Environ Microbiol Rep. 2014. PMID: 25646533 Review.
-
Polysaccharide lyases.Appl Biochem Biotechnol. 1986 Apr;12(2):135-76. doi: 10.1007/BF02798420. Appl Biochem Biotechnol. 1986. PMID: 3521491 Review.
Cited by
-
Immobilization of Purified Pectin Lyase from Pseudomonas putida onto Magnetic Lily Flowers (Lilium candidum L.) Nanoparticles and Applicability in Industrial Processes.Molecules. 2020 Jun 9;25(11):2671. doi: 10.3390/molecules25112671. Molecules. 2020. PMID: 32526868 Free PMC article.
-
Evolution and functional characterization of pectate lyase PEL12, a member of a highly expanded Clonostachys rosea polysaccharide lyase 1 family.BMC Microbiol. 2018 Nov 7;18(1):178. doi: 10.1186/s12866-018-1310-9. BMC Microbiol. 2018. PMID: 30404596 Free PMC article.
-
Research Progress on Phytopathogenic Fungi and Their Role as Biocontrol Agents.Front Microbiol. 2021 May 28;12:670135. doi: 10.3389/fmicb.2021.670135. eCollection 2021. Front Microbiol. 2021. PMID: 34122383 Free PMC article. Review.
-
Genome mining of Fusarium reveals structural and functional diversity of pectin lyases: a bioinformatics approach.3 Biotech. 2022 Oct;12(10):261. doi: 10.1007/s13205-022-03333-w. Epub 2022 Sep 6. 3 Biotech. 2022. PMID: 36082361 Free PMC article.
-
Differential Carbon Catabolite Repression and Hemicellulolytic Ability among Pathotypes of Colletotrichum lindemuthianum against Natural Plant Substrates.J Fungi (Basel). 2024 Jun 5;10(6):406. doi: 10.3390/jof10060406. J Fungi (Basel). 2024. PMID: 38921392 Free PMC article.
References
-
- Jayani RS, Saxena S, Gupta R. Microbial pectinolytic enzymes: a review. Process Biochem. 2005;40:2931–2944. doi: 10.1016/j.procbio.2005.03.026. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

