Identification of candidate genes for yeast engineering to improve bioethanol production in very high gravity and lignocellulosic biomass industrial fermentations
- PMID: 22152034
- PMCID: PMC3287136
- DOI: 10.1186/1754-6834-4-57
Identification of candidate genes for yeast engineering to improve bioethanol production in very high gravity and lignocellulosic biomass industrial fermentations
Abstract
Background: The optimization of industrial bioethanol production will depend on the rational design and manipulation of industrial strains to improve their robustness against the many stress factors affecting their performance during very high gravity (VHG) or lignocellulosic fermentations. In this study, a set of Saccharomyces cerevisiae genes found, through genome-wide screenings, to confer resistance to the simultaneous presence of different relevant stresses were identified as required for maximal fermentation performance under industrial conditions.
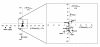
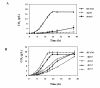
Results: Chemogenomics data were used to identify eight genes whose expression confers simultaneous resistance to high concentrations of glucose, acetic acid and ethanol, chemical stresses relevant for VHG fermentations; and eleven genes conferring simultaneous resistance to stresses relevant during lignocellulosic fermentations. These eleven genes were identified based on two different sets: one with five genes granting simultaneous resistance to ethanol, acetic acid and furfural, and the other with six genes providing simultaneous resistance to ethanol, acetic acid and vanillin. The expression of Bud31 and Hpr1 was found to lead to the increase of both ethanol yield and fermentation rate, while Pho85, Vrp1 and Ygl024w expression is required for maximal ethanol production in VHG fermentations. Five genes, Erg2, Prs3, Rav1, Rpb4 and Vma8, were found to contribute to the maintenance of cell viability in wheat straw hydrolysate and/or the maximal fermentation rate of this substrate.
Conclusions: The identified genes stand as preferential targets for genetic engineering manipulation in order to generate more robust industrial strains, able to cope with the most significant fermentation stresses and, thus, to increase ethanol production rate and final ethanol titers.
Figures
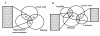

Similar articles
-
Genome-wide screening of Saccharomyces cerevisiae genes required to foster tolerance towards industrial wheat straw hydrolysates.J Ind Microbiol Biotechnol. 2014 Dec;41(12):1753-61. doi: 10.1007/s10295-014-1519-z. Epub 2014 Oct 7. J Ind Microbiol Biotechnol. 2014. PMID: 25287021
-
Contribution of PRS3, RPB4 and ZWF1 to the resistance of industrial Saccharomyces cerevisiae CCUG53310 and PE-2 strains to lignocellulosic hydrolysate-derived inhibitors.Bioresour Technol. 2015 Sep;191:7-16. doi: 10.1016/j.biortech.2015.05.006. Epub 2015 May 7. Bioresour Technol. 2015. PMID: 25974617
-
Robust industrial Saccharomyces cerevisiae strains for very high gravity bio-ethanol fermentations.J Biosci Bioeng. 2011 Aug;112(2):130-6. doi: 10.1016/j.jbiosc.2011.03.022. Epub 2011 May 2. J Biosci Bioeng. 2011. PMID: 21543257
-
Very high gravity (VHG) ethanolic brewing and fermentation: a research update.J Ind Microbiol Biotechnol. 2011 Sep;38(9):1133-44. doi: 10.1007/s10295-011-0999-3. Epub 2011 Jun 22. J Ind Microbiol Biotechnol. 2011. PMID: 21695540 Review.
-
Biotechnological strategies to overcome inhibitors in lignocellulose hydrolysates for ethanol production: review.Crit Rev Biotechnol. 2011 Mar;31(1):20-31. doi: 10.3109/07388551003757816. Epub 2010 May 31. Crit Rev Biotechnol. 2011. PMID: 20513164 Review.
Cited by
-
Increased expression of the yeast multidrug resistance ABC transporter Pdr18 leads to increased ethanol tolerance and ethanol production in high gravity alcoholic fermentation.Microb Cell Fact. 2012 Jul 27;11:98. doi: 10.1186/1475-2859-11-98. Microb Cell Fact. 2012. PMID: 22839110 Free PMC article.
-
Isolation of Saccharomyces Cerevisiae from Pineapple and Orange and Study of Metal's Effectiveness on Ethanol Production.Eur J Microbiol Immunol (Bp). 2017 Feb 27;7(1):76-91. doi: 10.1556/1886.2016.00035. eCollection 2017 Mar. Eur J Microbiol Immunol (Bp). 2017. PMID: 28386473 Free PMC article.
-
Dissecting a complex chemical stress: chemogenomic profiling of plant hydrolysates.Mol Syst Biol. 2013 Jun 18;9:674. doi: 10.1038/msb.2013.30. Mol Syst Biol. 2013. PMID: 23774757 Free PMC article.
-
Saccharomyces cerevisiae Cells Lacking the Zinc Vacuolar Transporter Zrt3 Display Improved Ethanol Productivity in Lignocellulosic Hydrolysates.J Fungi (Basel). 2022 Jan 14;8(1):78. doi: 10.3390/jof8010078. J Fungi (Basel). 2022. PMID: 35050019 Free PMC article.
-
High vanillin tolerance of an evolved Saccharomyces cerevisiae strain owing to its enhanced vanillin reduction and antioxidative capacity.J Ind Microbiol Biotechnol. 2014 Nov;41(11):1637-45. doi: 10.1007/s10295-014-1515-3. Epub 2014 Sep 28. J Ind Microbiol Biotechnol. 2014. PMID: 25261986
References
-
- Balat M, Balat H. Recent trends in global production and utilization of bio-ethanol fuel. Appl Energy. 2009;86:2273–2282. doi: 10.1016/j.apenergy.2009.03.015. - DOI
-
- Smeets EMW, Faaij APC, Lewandowski IM, Turkenburg WC. A bottom-up assessment and review of global bio-energy potentials to 2050. Progress in Energy and Combustion Science. 2007;33:56–106. doi: 10.1016/j.pecs.2006.08.001. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

