Genome-wide analytical approaches for reverse metabolic engineering of industrially relevant phenotypes in yeast
- PMID: 22152095
- PMCID: PMC3615171
- DOI: 10.1111/j.1567-1364.2011.00776.x
Genome-wide analytical approaches for reverse metabolic engineering of industrially relevant phenotypes in yeast
Abstract
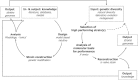
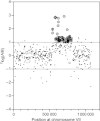
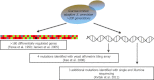
Successful reverse engineering of mutants that have been obtained by nontargeted strain improvement has long presented a major challenge in yeast biotechnology. This paper reviews the use of genome-wide approaches for analysis of Saccharomyces cerevisiae strains originating from evolutionary engineering or random mutagenesis. On the basis of an evaluation of the strengths and weaknesses of different methods, we conclude that for the initial identification of relevant genetic changes, whole genome sequencing is superior to other analytical techniques, such as transcriptome, metabolome, proteome, or array-based genome analysis. Key advantages of this technique over gene expression analysis include the independency of genome sequences on experimental context and the possibility to directly and precisely reproduce the identified changes in naive strains. The predictive value of genome-wide analysis of strains with industrially relevant characteristics can be further improved by classical genetics or simultaneous analysis of strains derived from parallel, independent strain improvement lineages.
© 2011 Federation of European Microbiological Societies. Published by Blackwell Publishing Ltd. All rights reserved.
Figures
Similar articles
-
Under pressure: evolutionary engineering of yeast strains for improved performance in fuels and chemicals production.Curr Opin Biotechnol. 2018 Apr;50:47-56. doi: 10.1016/j.copbio.2017.10.011. Epub 2017 Nov 20. Curr Opin Biotechnol. 2018. PMID: 29156423 Review.
-
Evolutionary engineering of Saccharomyces cerevisiae for improved industrially important properties.FEMS Yeast Res. 2012 Mar;12(2):171-82. doi: 10.1111/j.1567-1364.2011.00775.x. Epub 2011 Dec 23. FEMS Yeast Res. 2012. PMID: 22136139 Review.
-
Whole genome sequencing of Saccharomyces cerevisiae: from genotype to phenotype for improved metabolic engineering applications.BMC Genomics. 2010 Dec 22;11:723. doi: 10.1186/1471-2164-11-723. BMC Genomics. 2010. PMID: 21176163 Free PMC article.
-
Deciphering the Genic Basis of Yeast Fitness Variation by Simultaneous Forward and Reverse Genetics.Mol Biol Evol. 2017 Oct 1;34(10):2486-2502. doi: 10.1093/molbev/msx151. Mol Biol Evol. 2017. PMID: 28472365 Free PMC article.
-
Genome shuffling of Saccharomyces cerevisiae for enhanced glutathione yield and relative gene expression analysis using fluorescent quantitation reverse transcription polymerase chain reaction.J Microbiol Methods. 2016 Aug;127:188-192. doi: 10.1016/j.mimet.2016.06.012. Epub 2016 Jun 11. J Microbiol Methods. 2016. PMID: 27302037
Cited by
-
Nanopore sequencing enables near-complete de novo assembly of Saccharomyces cerevisiae reference strain CEN.PK113-7D.FEMS Yeast Res. 2017 Nov 1;17(7):fox074. doi: 10.1093/femsyr/fox074. FEMS Yeast Res. 2017. PMID: 28961779 Free PMC article.
-
Deconstructing the genetic basis of spent sulphite liquor tolerance using deep sequencing of genome-shuffled yeast.Biotechnol Biofuels. 2015 Mar 31;8:53. doi: 10.1186/s13068-015-0241-z. eCollection 2015. Biotechnol Biofuels. 2015. PMID: 25866561 Free PMC article.
-
Genome duplication and mutations in ACE2 cause multicellular, fast-sedimenting phenotypes in evolved Saccharomyces cerevisiae.Proc Natl Acad Sci U S A. 2013 Nov 5;110(45):E4223-31. doi: 10.1073/pnas.1305949110. Epub 2013 Oct 21. Proc Natl Acad Sci U S A. 2013. PMID: 24145419 Free PMC article.
-
Evolutionary Engineering in Chemostat Cultures for Improved Maltotriose Fermentation Kinetics in Saccharomyces pastorianus Lager Brewing Yeast.Front Microbiol. 2017 Sep 8;8:1690. doi: 10.3389/fmicb.2017.01690. eCollection 2017. Front Microbiol. 2017. PMID: 28943864 Free PMC article.
-
Saccharomyces cerevisiae strains for second-generation ethanol production: from academic exploration to industrial implementation.FEMS Yeast Res. 2017 Aug 1;17(5):fox044. doi: 10.1093/femsyr/fox044. FEMS Yeast Res. 2017. PMID: 28899031 Free PMC article. Review.
References
-
- Abbott DA, Knijnenburg TA, de Poorter LMI, Reinders MJT, Pronk JT, van Maris AJA. Generic and specific transcriptional responses to different weak organic acids in anaerobic chemostat cultures of Saccharomyces cerevisiae. FEMS Yeast Res. 2007;7:819–833. - PubMed
-
- Abbott DA, Zelle RM, Pronk JT, van Maris AJA. Metabolic engineering of Saccharomyces cerevisiae for production of carboxylic acids: current status and challenges. FEMS Yeast Res. 2009;9:1123–1136. - PubMed
-
- Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. - PubMed
-
- Anderson JC, Dueber JE, Leguia M, Wu GC, Goler JA, Arkin AP, Keasling JD. BglBricks: a flexible standard for biological part assembly. J Biol Eng. 2010;4:1. DOI: 10.1186/1754-1611-4-1. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

