Detailed genetic analysis of hemagglutinin-neuraminidase glycoprotein gene in human parainfluenza virus type 1 isolates from patients with acute respiratory infection between 2002 and 2009 in Yamagata prefecture, Japan
- PMID: 22152158
- PMCID: PMC3295729
- DOI: 10.1186/1743-422X-8-533
Detailed genetic analysis of hemagglutinin-neuraminidase glycoprotein gene in human parainfluenza virus type 1 isolates from patients with acute respiratory infection between 2002 and 2009 in Yamagata prefecture, Japan
Abstract
Background: Human parainfluenza virus type 1 (HPIV1) causes various acute respiratory infections (ARI). Hemagglutinin-neuraminidase (HN) glycoprotein of HPIV1 is a major antigen. However, the molecular epidemiology and genetic characteristics of such ARI are not exactly known. Recent studies suggested that a phylogenetic analysis tool, namely the maximum likelihood (ML) method, may be applied to estimate the evolutionary time scale of various viruses. Thus, we conducted detailed genetic analyses including homology analysis, phylogenetic analysis (using both the neighbor joining (NJ) and ML methods), and analysis of the pairwise distances of HN gene in HPIV1 isolated from patients with ARI in Yamagata prefecture, Japan.
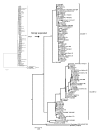
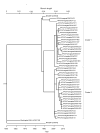
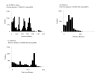
Results: A few substitutions of nucleotides in the second binding site of HN gene were observed among the present isolates. The strains were classified into two major clusters in the phylogenetic tree by the NJ method. Another phylogenetic tree constructed by the ML method showed that the strains diversified in the late 1980s. No positively selected sites were found in the present strains. Moreover, the pairwise distance among the present isolates was relatively short.
Conclusions: The evolution of HN gene in the present HPIV1 isolates was relatively slow. The ML method may be a useful phylogenetic method to estimate the evolutionary time scale of HPIV and other viruses.
Figures
Similar articles
-
Genetic analyses of the fusion protein genes in human parainfluenza virus types 1 and 3 among patients with acute respiratory infections in Eastern Japan from 2011 to 2015.J Med Microbiol. 2017 Feb;66(2):160-168. doi: 10.1099/jmm.0.000431. J Med Microbiol. 2017. PMID: 28266286
-
Molecular evolution of the haemagglutinin-neuraminidase gene in human parainfluenza virus type 3 isolates from children with acute respiratory illness in Yamagata prefecture, Japan.J Med Microbiol. 2014 Apr;63(Pt 4):570-577. doi: 10.1099/jmm.0.068189-0. Epub 2014 Jan 25. J Med Microbiol. 2014. PMID: 24464692
-
Genetic characteristics of human parainfluenza viruses 1-4 associated with acute lower respiratory tract infection in Chinese children, during 2015-2021.Microbiol Spectr. 2024 Oct 3;12(10):e0343223. doi: 10.1128/spectrum.03432-23. Epub 2024 Sep 12. Microbiol Spectr. 2024. PMID: 39264196 Free PMC article.
-
Detailed genetic analyses of the HN gene in human respirovirus 3 detected in children with acute respiratory illness in the Iwate Prefecture, Japan.Infect Genet Evol. 2018 Apr;59:155-162. doi: 10.1016/j.meegid.2018.01.021. Epub 2018 Feb 14. Infect Genet Evol. 2018. PMID: 29408530
-
New antiviral approaches for human parainfluenza: Inhibiting the haemagglutinin-neuraminidase.Antiviral Res. 2019 Jul;167:89-97. doi: 10.1016/j.antiviral.2019.04.001. Epub 2019 Apr 3. Antiviral Res. 2019. PMID: 30951732 Review.
Cited by
-
Molecular Evolution and Characterization of Hemagglutinin (H) in Peste des Petits Ruminants Virus.PLoS One. 2016 Apr 1;11(4):e0152587. doi: 10.1371/journal.pone.0152587. eCollection 2016. PLoS One. 2016. PMID: 27035347 Free PMC article.
-
Genetic diversity and epidemiology of human rhinovirus among children with severe acute respiratory tract infection in Guangzhou, China.Virol J. 2021 Aug 23;18(1):174. doi: 10.1186/s12985-021-01645-6. Virol J. 2021. PMID: 34425845 Free PMC article.
-
Development of Primer Panels for Amplicon Sequencing of Human Parainfluenza Viruses Type 1 and 2.Int J Mol Sci. 2024 Dec 6;25(23):13119. doi: 10.3390/ijms252313119. Int J Mol Sci. 2024. PMID: 39684830 Free PMC article.
-
Human parainfluenza virus type 3 (HPIV3) induces production of IFNγ and RANTES in human nasal epithelial cells (HNECs).J Inflamm (Lond). 2015 Feb 21;12:16. doi: 10.1186/s12950-015-0054-7. eCollection 2015. J Inflamm (Lond). 2015. PMID: 25722655 Free PMC article.
-
Molecular characterization of human parainfluenza virus type 1 in infants attending Mbagathi District Hospital, Nairobi, Kenya: a retrospective study.Virus Genes. 2013 Dec;47(3):439-47. doi: 10.1007/s11262-013-0970-7. Epub 2013 Aug 17. Virus Genes. 2013. PMID: 23955068
References
-
- Karron RA, Collins PL. In: Fields virology. 5. Knipe DM, Howley PM, editor. Vol. 1. Philadelphia: Lippincott Williams & Wilkins; 2007. Parainfluenza viruses; pp. 1497–1526.
-
- Griffin MR, Walker FJ, Iwane MK, Weinberg GA, Staat MA, Erdman DD. New Vaccine Surveillance Network Study Group. Epidemiology of respiratory infections in young children: insights from the new vaccine surveillance network. Pediatr Infect Dis J. 2004;23(11 Suppl):S188–192. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

