Regulation of ribonucleotide reductase in response to iron deficiency
- PMID: 22152479
- PMCID: PMC3240860
- DOI: 10.1016/j.molcel.2011.09.021
Regulation of ribonucleotide reductase in response to iron deficiency
Abstract
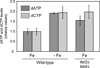
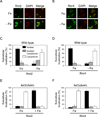
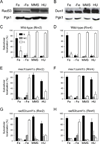
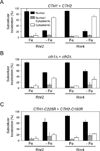
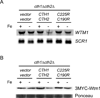
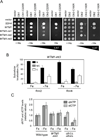
Ribonucleotide reductase (RNR) is an essential enzyme required for DNA synthesis and repair. Although iron is necessary for class Ia RNR activity, little is known about the mechanisms that control RNR in response to iron deficiency. In this work, we demonstrate that yeast cells control RNR function during iron deficiency by redistributing the Rnr2-Rnr4 small subunit from the nucleus to the cytoplasm. Our data support a Mec1/Rad53-independent mechanism in which the iron-regulated Cth1/Cth2 mRNA-binding proteins specifically interact with the WTM1 mRNA in response to iron scarcity and promote its degradation. The resulting decrease in the nuclear-anchoring Wtm1 protein levels leads to the redistribution of the Rnr2-Rnr4 heterodimer to the cytoplasm, where it assembles as an active RNR complex and increases deoxyribonucleoside triphosphate levels. When iron is scarce, yeast selectively optimizes RNR function at the expense of other non-essential iron-dependent processes that are repressed, to allow DNA synthesis and repair.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

Comment in
-
Regulation of ribonucleotide reductase during iron limitation.Mol Cell. 2011 Dec 9;44(5):683-4. doi: 10.1016/j.molcel.2011.11.011. Mol Cell. 2011. PMID: 22152471 Free PMC article.
Similar articles
-
Yeast Dun1 Kinase Regulates Ribonucleotide Reductase Small Subunit Localization in Response to Iron Deficiency.J Biol Chem. 2016 Apr 29;291(18):9807-17. doi: 10.1074/jbc.M116.720862. Epub 2016 Mar 12. J Biol Chem. 2016. PMID: 26970775 Free PMC article.
-
Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1.Genes Dev. 2006 Feb 1;20(3):334-44. doi: 10.1101/gad.1380506. Genes Dev. 2006. PMID: 16452505 Free PMC article.
-
Yeast Dun1 kinase regulates ribonucleotide reductase inhibitor Sml1 in response to iron deficiency.Mol Cell Biol. 2014 Sep;34(17):3259-71. doi: 10.1128/MCB.00472-14. Epub 2014 Jun 23. Mol Cell Biol. 2014. PMID: 24958100 Free PMC article.
-
Post-transcriptional regulation of iron homeostasis in Saccharomyces cerevisiae.Int J Mol Sci. 2013 Jul 30;14(8):15785-809. doi: 10.3390/ijms140815785. Int J Mol Sci. 2013. PMID: 23903042 Free PMC article. Review.
-
Function and regulation of yeast ribonucleotide reductase: cell cycle, genotoxic stress, and iron bioavailability.Biomed J. 2013 Mar-Apr;36(2):51-8. doi: 10.4103/2319-4170.110398. Biomed J. 2013. PMID: 23644233 Review.
Cited by
-
A comprehensive mechanistic model of iron metabolism in Saccharomyces cerevisiae.Metallomics. 2019 Nov 1;11(11):1779-1799. doi: 10.1039/c9mt00199a. Epub 2019 Sep 18. Metallomics. 2019. PMID: 31531508 Free PMC article.
-
Regulatory and pathogenic mechanisms in response to iron deficiency and excess in fungi.Microb Biotechnol. 2023 Nov;16(11):2053-2071. doi: 10.1111/1751-7915.14346. Epub 2023 Oct 7. Microb Biotechnol. 2023. PMID: 37804207 Free PMC article. Review.
-
Yeast Cth2 protein represses the translation of ARE-containing mRNAs in response to iron deficiency.PLoS Genet. 2018 Jun 18;14(6):e1007476. doi: 10.1371/journal.pgen.1007476. eCollection 2018 Jun. PLoS Genet. 2018. PMID: 29912874 Free PMC article.
-
mRNA-binding protein tristetraprolin is essential for cardiac response to iron deficiency by regulating mitochondrial function.Proc Natl Acad Sci U S A. 2018 Jul 3;115(27):E6291-E6300. doi: 10.1073/pnas.1804701115. Epub 2018 Jun 18. Proc Natl Acad Sci U S A. 2018. PMID: 29915044 Free PMC article.
-
Regulation of ribonucleotide reductase during iron limitation.Mol Cell. 2011 Dec 9;44(5):683-4. doi: 10.1016/j.molcel.2011.11.011. Mol Cell. 2011. PMID: 22152471 Free PMC article.
References
-
- Cavanaugh PF, Jr, Porter CW, Tukalo D, Frankfurt OS, Pavelic ZP, Bergeron RJ. Characterization of L1210 cell growth inhibition by the bacterial iron chelators parabactin and compound II. Cancer Res. 1985;45:4754–4759. - PubMed
-
- Chabes A, Domkin V, Thelander L. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J Biol Chem. 1999;274:36679–36683. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

