An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis
- PMID: 22153071
- PMCID: PMC3251960
- DOI: 10.1016/j.cell.2011.10.043
An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis
Erratum in
- Cell. 2013 Feb 28;152(5):1196
Abstract
Hepatocyte nuclear factor 4α (HNF4α) is essential for liver development and hepatocyte function. Here, we show that transient inhibition of HNF4α initiates hepatocellular transformation through a microRNA-inflammatory feedback loop circuit consisting of miR-124, IL6R, STAT3, miR-24, and miR-629. Moreover, we show that, once this circuit is activated, it maintains suppression of HNF4α and sustains oncogenesis. Systemic administration of miR-124, which modulates inflammatory signaling, prevents and suppresses hepatocellular carcinogenesis by inducing tumor-specific apoptosis without toxic side effects. As we also show that this HNF4α circuit is perturbed in human hepatocellular carcinomas, our data raise the possibility that manipulation of this microRNA feedback-inflammatory loop has therapeutic potential for treating liver cancer.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
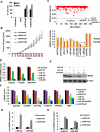
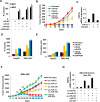
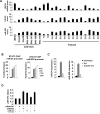
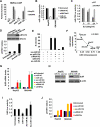

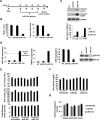
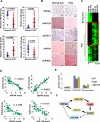
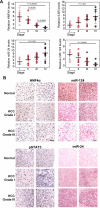
Comment in
-
How transient becomes stable: an epigenetic switch linking liver inflammation and tumorigenesis.J Hepatol. 2012 Oct;57(4):910-2. doi: 10.1016/j.jhep.2012.05.017. Epub 2012 Jun 2. J Hepatol. 2012. PMID: 22668642 No abstract available.
-
An inflammatory proposal for hepatocarcinogenesis.Hepatology. 2012 Jul;56(1):382-4. doi: 10.1002/hep.25759. Hepatology. 2012. PMID: 22876364 No abstract available.
References
-
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr. Stat3 as an oncogene. Cell. 1999;98:295–303. - PubMed
-
- Calvisi DF, Ladu S, Gorden A, Farina M, Conner EA, Lee JS, Factor VM, Thorgeirsson SS. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006;130:1117–1128. - PubMed
-
- Cattin AL, Le Beyec J, Barreau F, Saint-Just S, Houllier A, Gonzalez FJ, Robine S, Pincon-Raymond M, Cardot P, Lacasa M, et al. Hepatocyte nuclear factor 4alpha, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol Cell Biol. 2009;29:6294–6308. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

