Structural and dynamic determinants of protein-peptide recognition
- PMID: 22153506
- PMCID: PMC3240807
- DOI: 10.1016/j.str.2011.09.014
Structural and dynamic determinants of protein-peptide recognition
Abstract
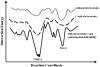
Protein-peptide interactions play important roles in many cellular processes, including signal transduction, trafficking, and immune recognition. Protein conformational changes upon binding, an ill-defined peptide binding surface, and the large number of peptide degrees of freedom make the prediction of protein-peptide interactions particularly challenging. To address these challenges, we perform rapid molecular dynamics simulations in order to examine the energetic and dynamic aspects of protein-peptide binding. We find that, in most cases, we recapitulate the native binding sites and native-like poses of protein-peptide complexes. Inclusion of electrostatic interactions in simulations significantly improves the prediction accuracy. Our results also highlight the importance of protein conformational flexibility, especially side-chain movement, which allows the peptide to optimize its conformation. Our findings not only demonstrate the importance of sufficient sampling of the protein and peptide conformations, but also reveal the possible effects of electrostatics and conformational flexibility on peptide recognition.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures


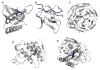
Similar articles
-
Indirect readout in protein-peptide recognition: a different story from classical biomolecular recognition.J Chem Inf Model. 2014 Jul 28;54(7):2022-32. doi: 10.1021/ci5000246. Epub 2014 Jul 16. J Chem Inf Model. 2014. PMID: 24999015
-
A protocol for CABS-dock protein-peptide docking driven by side-chain contact information.Biomed Eng Online. 2017 Aug 18;16(Suppl 1):73. doi: 10.1186/s12938-017-0363-6. Biomed Eng Online. 2017. PMID: 28830545 Free PMC article.
-
A unified conformational selection and induced fit approach to protein-peptide docking.PLoS One. 2013;8(3):e58769. doi: 10.1371/journal.pone.0058769. Epub 2013 Mar 13. PLoS One. 2013. PMID: 23516555 Free PMC article.
-
Modeling of Protein Structural Flexibility and Large-Scale Dynamics: Coarse-Grained Simulations and Elastic Network Models.Int J Mol Sci. 2018 Nov 6;19(11):3496. doi: 10.3390/ijms19113496. Int J Mol Sci. 2018. PMID: 30404229 Free PMC article. Review.
-
Understanding the challenges of protein flexibility in drug design.Expert Opin Drug Discov. 2015 Dec;10(12):1301-13. doi: 10.1517/17460441.2015.1094458. Epub 2015 Sep 28. Expert Opin Drug Discov. 2015. PMID: 26414598 Review.
Cited by
-
Mutagenesis of DsbAss is Crucial for the Signal Recognition Particle Mechanism in Escherichia coli: Insights from Molecular Dynamics Simulations.Biomolecules. 2019 Apr 3;9(4):133. doi: 10.3390/biom9040133. Biomolecules. 2019. PMID: 30987187 Free PMC article.
-
Sensory plasticity caused by up-down regulation encodes the information of short-term learning and memory.iScience. 2025 Mar 13;28(4):112215. doi: 10.1016/j.isci.2025.112215. eCollection 2025 Apr 18. iScience. 2025. PMID: 40224011 Free PMC article.
-
Leveraging RAS-mSIN1 interaction to selectively inhibit mTORC2 employing competitive RAS binding peptide: implications in breast cancer metastasis.Oncogene. 2025 Aug 23. doi: 10.1038/s41388-025-03516-8. Online ahead of print. Oncogene. 2025. PMID: 40847128
-
Leveraging Therapeutic Proteins and Peptides from Lumbricus Earthworms: Targeting SOCS2 E3 Ligase for Cardiovascular Therapy through Molecular Dynamics Simulations.Int J Mol Sci. 2024 Oct 8;25(19):10818. doi: 10.3390/ijms251910818. Int J Mol Sci. 2024. PMID: 39409145 Free PMC article.
-
Applications of Discrete Molecular Dynamics in biology and medicine.Curr Opin Struct Biol. 2016 Apr;37:9-13. doi: 10.1016/j.sbi.2015.11.001. Epub 2015 Nov 28. Curr Opin Struct Biol. 2016. PMID: 26638022 Free PMC article. Review.
References
-
- Aita T, Nishigaki K, Husimi Y. Toward the fast blind docking of a peptide to a target protein by using a four-body statistical pseudo-potential. Comput Biol Chem. 2010;34:53–62. - PubMed
-
- Anderson AC, O’Neil RH, Surti TS, Stroud RM. Approaches to solving the rigid receptor problem by identifying a minimal set of flexible residues during ligand docking. Chem Biol. 2001;8:445–457. - PubMed
-
- Antes I. DynaDock: A new molecular dynamics-based algorithm for protein-peptide docking including receptor flexibility. Proteins. 2010;78:1084–1104. - PubMed
-
- Birdsall B, Feeney J, Tendler SJ, Hammond SJ, Roberts GC. Dihydrofolate reductase: multiple conformations and alternative modes of substrate binding. Biochemistry. 1989;28:2297–2305. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

