CRABP-II methylation: a critical determinant of retinoic acid resistance of medulloblastoma cells
- PMID: 22153617
- PMCID: PMC5528385
- DOI: 10.1016/j.molonc.2011.11.004
CRABP-II methylation: a critical determinant of retinoic acid resistance of medulloblastoma cells
Abstract
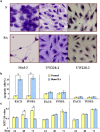
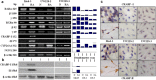
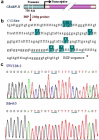
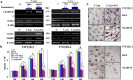
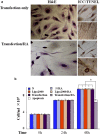
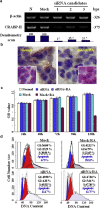
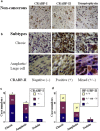
Medulloblastoma cells exhibit varied responses to therapy by all-trans retinoic acid (RA). The underlying mechanism for such diverse effects however remains largely unclear. In this study, we attempted to elucidate the molecular basis of RA resistance through the study of RA signaling components in both RA-sensitive (Med-3) and RA-resistant (UW228-2 and UW228-3) medulloblastoma cells. The results revealed that RARα/β/γ and RXRα/β/γ were found in the three cell lines. Expression of CRABP-I and CRABP-II was seen in Med-3 cells, up-regulated when treated with RA, but was absent in UW228-2 and UW228-3 cells regardless of RA treatment. Bisulfite sequencing revealed 8 methylated CG sites at the promoter region of CRABP-II in UW228-2 and UW228-3 but not in Med-3 cells. Demethylation by 5-aza-2'-deoxycytidine recovered CRABP-II expression. Upon restoration of CRABP-II expression, both UW228-2 and UW228-3 cells responded to RA treatment by forming neuronal-like differentiation, synaptophysin expression, β-III tubulin upregulation, and apoptosis. Furthermore, CRABP-II specific siRNA reduced RA sensitivity in Med-3 cells. Tissue microarray-based immunohistochemical staining showed variable CRABP-II expression patterns among 104 medulloblastoma cases, ranging from negative (42.3%), partly positive (14.4%) to positive (43.3%). CRABP-II expression was positively correlated with synaptophysin (rs = 0.317; p = 0.001) but not with CRABP-I expression (p > 0.05). In conclusion, aberrant methylation in CRABP-II reduces the expression of CRABP-II that in turn confers RA resistance in medulloblastoma cells. Determination of CRABP-II expression or methylation status may enable a personalized RA therapy in patients with medulloblastomas and other types of cancers.
Copyright © 2011 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures
References
-
- Abu-Abed, S. , MacLean, G. , Fraulob, V. , Chambon, P. , Petkovich, M. , Dollé, P. , 2002. Differential expression of the retinoic acid-metabolizing enzymes CYP26A1 and CYP26B1 during murine organogenesis. Mech. Dev.. 110, 173–177. - PubMed
-
- Aström, A. , Tavakkol, A. , Pettersson, U. , Cromie, M. , Elder, J.T. , Voorhees, J.J. , 1991. Molecular cloning of two human cellular retinoic acid-binding proteins (CRABP). Retinoic acid-induced expression of CRABP-II but not CRABP-I in adult human skin in vivo and in skin fibroblasts in vitro. J. Biol. Chem.. 266, 17662–17666. - PubMed
-
- Bergamaschi, D. , Gasco, M. , Hiller, L. , Sullivan, A. , Syed, N. , Trigiante, G. , Yulug, I. , Merlano, M. , Numico, G. , Comino, A. , Attard, M. , Reelfs, O. , 2003. P53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell.. 3, 387–402. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

