Deconstructing p53 transcriptional networks in tumor suppression
- PMID: 22154076
- PMCID: PMC3541866
- DOI: 10.1016/j.tcb.2011.10.006
Deconstructing p53 transcriptional networks in tumor suppression
Abstract
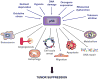
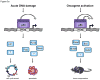
p53 is a pivotal tumor suppressor that induces apoptosis, cell-cycle arrest and senescence in response to stress signals. Although p53 transcriptional activation is important for these responses, the mechanisms underlying tumor suppression have been elusive. To date, no single or compound mouse knockout of specific p53 target genes has recapitulated the dramatic tumor predisposition that characterizes p53-null mice. Recently, however, analysis of knock-in mice expressing p53 transactivation domain mutants has revealed a group of primarily novel direct p53 target genes that may mediate tumor suppression in vivo. We present here an overview of well-known p53 target genes and the tumor phenotypes of the cognate knockout mice, and address the recent identification of new p53 transcriptional targets and how they enhance our understanding of p53 transcriptional networks central for tumor suppression.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures


References
-
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. - PubMed
-
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316– 323. - PubMed
-
- Halazonetis TD, et al. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. - PubMed
-
- Christophorou MA, et al. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006;443:214–217. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

