Directed evolution to re-adapt a co-evolved network within an enzyme
- PMID: 22154561
- PMCID: PMC3657141
- DOI: 10.1016/j.jbiotec.2011.11.017
Directed evolution to re-adapt a co-evolved network within an enzyme
Abstract
We have previously used targeted active-site saturation mutagenesis to identify a number of transketolase single mutants that improved activity towards either glycolaldehyde (GA), or the non-natural substrate propionaldehyde (PA). Here, all attempts to recombine the singles into double mutants led to unexpected losses of specific activity towards both substrates. A typical trade-off occurred between soluble expression levels and specific activity for all single mutants, but many double mutants decreased both properties more severely suggesting a critical loss of protein stability or native folding. Statistical coupling analysis (SCA) of a large multiple sequence alignment revealed a network of nine co-evolved residues that affected all but one double mutant. Such networks maintain important functional properties such as activity, specificity, folding, stability, and solubility and may be rapidly disrupted by introducing one or more non-naturally occurring mutations. To identify variants of this network that would accept and improve upon our best D469 mutants for activity towards PA, we created a library of random single, double and triple mutants across seven of the co-evolved residues, combining our D469 variants with only naturally occurring mutations at the remaining sites. A triple mutant cluster at D469, E498 and R520 was found to behave synergistically for the specific activity towards PA. Protein expression was severely reduced by E498D and improved by R520Q, yet variants containing both mutations led to improved specific activity and enzyme expression, but with loss of solubility and the formation of inclusion bodies. D469S and R520Q combined synergistically to improve k(cat) 20-fold for PA, more than for any previous transketolase mutant. R520Q also doubled the specific activity of the previously identified D469T to create our most active transketolase mutant to date. Our results show that recombining active-site mutants obtained by saturation mutagenesis can rapidly destabilise critical networks of co-evolved residues, whereas beneficial single mutants can be retained and improved upon by randomly recombining them with natural variants at other positions in the network.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures

 ), and the initially created double mutants obtained by recombining the singles (□). Double and triple mutants obtained from the new library guided by SCA, and also for D469T/R520Q, are shown as triangles (▵) and interpolated with a dashed line.
), and the initially created double mutants obtained by recombining the singles (□). Double and triple mutants obtained from the new library guided by SCA, and also for D469T/R520Q, are shown as triangles (▵) and interpolated with a dashed line.
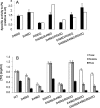
 ) soluble fraction and (■) final purified enzyme concentrations.
) soluble fraction and (■) final purified enzyme concentrations.

Similar articles
-
Directed evolution of transketolase substrate specificity towards an aliphatic aldehyde.J Biotechnol. 2008 Apr 30;134(3-4):240-5. doi: 10.1016/j.jbiotec.2008.01.018. Epub 2008 Feb 13. J Biotechnol. 2008. PMID: 18342970
-
Structural Analysis of an Evolved Transketolase Reveals Divergent Binding Modes.Sci Rep. 2016 Oct 21;6:35716. doi: 10.1038/srep35716. Sci Rep. 2016. PMID: 27767080 Free PMC article.
-
Second generation engineering of transketolase for polar aromatic aldehyde substrates.Enzyme Microb Technol. 2015 Apr;71:45-52. doi: 10.1016/j.enzmictec.2015.01.008. Epub 2015 Jan 31. Enzyme Microb Technol. 2015. PMID: 25765309
-
Crystallography and mutagenesis of transketolase: mechanistic implications for enzymatic thiamin catalysis.Biochim Biophys Acta. 1998 Jun 29;1385(2):387-98. doi: 10.1016/s0167-4838(98)00082-x. Biochim Biophys Acta. 1998. PMID: 9655943 Review.
-
Laboratory-directed protein evolution.Microbiol Mol Biol Rev. 2005 Sep;69(3):373-92. doi: 10.1128/MMBR.69.3.373-392.2005. Microbiol Mol Biol Rev. 2005. PMID: 16148303 Free PMC article. Review.
Cited by
-
Biophysical characterization of the inactivation of E. coli transketolase by aqueous co-solvents.Sci Rep. 2021 Dec 8;11(1):23584. doi: 10.1038/s41598-021-03001-8. Sci Rep. 2021. PMID: 34880340 Free PMC article.
-
Directed evolution for soluble and active periplasmic expression of bovine enterokinase in Escherichia coli.Sci Rep. 2022 Oct 21;12(1):17721. doi: 10.1038/s41598-022-22574-6. Sci Rep. 2022. PMID: 36271247 Free PMC article.
-
Enzyme stabilisation due to incorporation of a fluorinated non-natural amino acid at the protein surface.Sci Rep. 2024 Nov 14;14(1):28080. doi: 10.1038/s41598-024-79711-6. Sci Rep. 2024. PMID: 39543195 Free PMC article.
-
Therapeutic enzyme engineering using a generative neural network.Sci Rep. 2022 Jan 27;12(1):1536. doi: 10.1038/s41598-022-05195-x. Sci Rep. 2022. PMID: 35087131 Free PMC article.
-
Correlated positions in protein evolution and engineering.J Ind Microbiol Biotechnol. 2017 May;44(4-5):687-695. doi: 10.1007/s10295-016-1811-1. Epub 2016 Aug 11. J Ind Microbiol Biotechnol. 2017. PMID: 27514664 Review.
References
-
- Albery W.J., Knowles J.R. Evolution of enzyme function and the development of catalytic efficiency. Biochemistry. 1976;15:5631–5640. - PubMed
-
- Aucamp J.P., Martinez-Torres R.J., Hibbert E.G., Dalby P.A. A microplate-based evaluation of complex denaturation pathways: structural stability of Escherichia coli transketolase. Biotechnol. Bioeng. 2008;99:1303–1310. - PubMed
-
- Beadle B.M., Shoichet B.K. Structural bases of stability-function tradeoffs in enzymes. J. Mol. Biol. 2002;321:285–296. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

