Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14
- PMID: 22155632
- PMCID: PMC3295388
- DOI: 10.1093/jxb/err388
Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14
Abstract
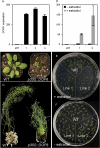
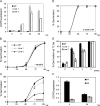
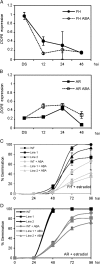
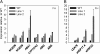
Seed dormancy prevents seeds from germinating under environmental conditions unfavourable for plant growth and development and constitutes an evolutionary advantage. Dry storage, also known as after-ripening, gradually decreases seed dormancy by mechanisms not well understood. An Arabidopsis thaliana DOF transcription factor gene (DOF6) affecting seed germination has been characterized. The transcript levels of this gene accumulate in dry seeds and decay gradually during after-ripening and also upon seed imbibition. While constitutive over-expression of DOF6 produced aberrant growth and sterility in the plant, its over-expression induced upon seed imbibition triggered delayed germination, abscisic acid (ABA)-hypersensitive phenotypes and increased expression of the ABA biosynthetic gene ABA1 and ABA-related stress genes. Wild-type germination and gene expression were gradually restored during seed after-ripening, despite of DOF6-induced over-expression. DOF6 was found to interact in a yeast two-hybrid system and in planta with TCP14, a previously described positive regulator of seed germination. The expression of ABA1 and ABA-related stress genes was also enhanced in tcp14 knock-out mutants. Taken together, these results indicate that DOF6 negatively affects seed germination and opposes TCP14 function in the regulation of a specific set of ABA-related genes.
Figures


Similar articles
-
CHOTTO1, a putative double APETALA2 repeat transcription factor, is involved in abscisic acid-mediated repression of gibberellin biosynthesis during seed germination in Arabidopsis.Plant Physiol. 2009 Oct;151(2):641-54. doi: 10.1104/pp.109.142018. Epub 2009 Jul 31. Plant Physiol. 2009. PMID: 19648230 Free PMC article.
-
The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds.Plant Cell. 2012 Jul;24(7):2826-38. doi: 10.1105/tpc.112.100214. Epub 2012 Jul 24. Plant Cell. 2012. PMID: 22829147 Free PMC article.
-
The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development.Plant J. 2016 Feb;85(4):451-65. doi: 10.1111/tpj.13118. Epub 2016 Feb 5. Plant J. 2016. PMID: 26729600
-
From the Outside to the Inside: New Insights on the Main Factors That Guide Seed Dormancy and Germination.Genes (Basel). 2020 Dec 31;12(1):52. doi: 10.3390/genes12010052. Genes (Basel). 2020. PMID: 33396410 Free PMC article. Review.
-
Parental and Environmental Control of Seed Dormancy in Arabidopsis thaliana.Annu Rev Plant Biol. 2022 May 20;73:355-378. doi: 10.1146/annurev-arplant-102820-090750. Epub 2022 Feb 9. Annu Rev Plant Biol. 2022. PMID: 35138879 Review.
Cited by
-
The family of DOF transcription factors in Brachypodium distachyon: phylogenetic comparison with rice and barley DOFs and expression profiling.BMC Plant Biol. 2012 Nov 5;12:202. doi: 10.1186/1471-2229-12-202. BMC Plant Biol. 2012. PMID: 23126376 Free PMC article.
-
Genome-wide identification and functional analysis of Dof transcription factor family in Camelina sativa.BMC Genomics. 2022 Dec 8;23(1):812. doi: 10.1186/s12864-022-09056-9. BMC Genomics. 2022. PMID: 36476342 Free PMC article.
-
Transcriptome analyses of the Dof-like gene family in grapevine reveal its involvement in berry, flower and seed development.Hortic Res. 2016 Aug 31;3:16042. doi: 10.1038/hortres.2016.42. eCollection 2016. Hortic Res. 2016. PMID: 27610237 Free PMC article.
-
DOF transcription factors: Specific regulators of plant biological processes.Front Plant Sci. 2023 Jan 20;14:1044918. doi: 10.3389/fpls.2023.1044918. eCollection 2023. Front Plant Sci. 2023. PMID: 36743498 Free PMC article. Review.
-
Genome-Wide Identification, Characterization, and Transcript Analysis of the TCP Transcription Factors in Vitis vinifera.Front Genet. 2019 Dec 20;10:1276. doi: 10.3389/fgene.2019.01276. eCollection 2019. Front Genet. 2019. PMID: 31921312 Free PMC article.
References
-
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. - PubMed
-
- Audran C, Liotenberg S, Gonneau M, North H, Frey A, Tap-Waksman K, Vartanian N, Marion-Poll A. Localisation and expression of zeaxanthin epoxidase mRNA in Arabidopsis in response to drough stress and during seed development. Australian Journal of Plant Physiology. 2001;28:1161–1173.
-
- Barrero JM, Millar AA, Griffiths J, Czechowski T, Scheible WR, Udvardi M, Reid JB, Ross JJ, Jacobsen JV, Gubler F. Gene expression profiling identifies two regulatory genes controlling dormancy and ABA sensitivity in Arabidopsis seeds. The Plant Journal. 2010;61:611–622. - PubMed
-
- Bove J, Lucas P, Godin B, Oge L, Jullien M, Grappin P. Gene expression analysis by cDNA-AFLP highlights a set of new signaling networks and translational control during seed dormancy breaking in Nicotiana plumbaginifolia. Plant Molecular Biology. 2005;57:593–612. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

