Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5
- PMID: 22156057
- PMCID: PMC3326321
- DOI: 10.1093/nar/gkr1192
Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5
Abstract
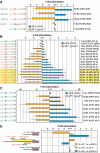
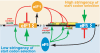
An AUG in an optimal nucleotide context is the preferred translation initiation site in eukaryotic cells. Interactions among translation initiation factors, including eIF1 and eIF5, govern start codon selection. Experiments described here showed that high intracellular eIF5 levels reduced the stringency of start codon selection in human cells. In contrast, high intracellular eIF1 levels increased stringency. High levels of eIF5 induced translation of inhibitory upstream open reading frames (uORFs) in eIF5 mRNA that initiate with AUG codons in conserved poor contexts. This resulted in reduced translation from the downstream eIF5 start codon, indicating that eIF5 autoregulates its own synthesis. As with eIF1, which is also autoregulated through translation initiation, features contributing to eIF5 autoregulation show deep evolutionary conservation. The results obtained provide the basis for a model in which auto- and cross-regulation of eIF5 and eIF1 translation establish a regulatory feedback loop that would stabilize the stringency of start codon selection.
Figures


References
-
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

