Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing
- PMID: 22156058
- PMCID: PMC3287169
- DOI: 10.1093/nar/gkr1146
Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing
Abstract
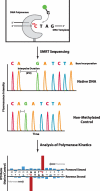
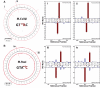
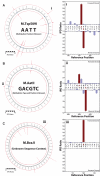
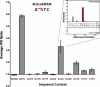
DNA methylation is the most common form of DNA modification in prokaryotic and eukaryotic genomes. We have applied the method of single-molecule, real-time (SMRT®) DNA sequencing that is capable of direct detection of modified bases at single-nucleotide resolution to characterize the specificity of several bacterial DNA methyltransferases (MTases). In addition to previously described SMRT sequencing of N6-methyladenine and 5-methylcytosine, we show that N4-methylcytosine also has a specific kinetic signature and is therefore identifiable using this approach. We demonstrate for all three prokaryotic methylation types that SMRT sequencing confirms the identity and position of the methylated base in cases where the MTase specificity was previously established by other methods. We then applied the method to determine the sequence context and methylated base identity for three MTases with unknown specificities. In addition, we also find evidence of unanticipated MTase promiscuity with some enzymes apparently also modifying sequences that are related, but not identical, to the cognate site.
Figures


References
-
- Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 1995;253:618–632. - PubMed
-
- Wu JC, Santi DV. Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem. 1987;262:4778–4786. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

