Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation
- PMID: 22156206
- PMCID: PMC3243055
- DOI: 10.1101/gad.179184.111
Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation
Abstract
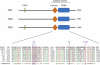
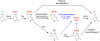
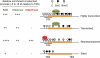
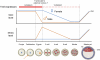
Ten-eleven translocation 1-3 (Tet1-3) proteins have recently been discovered in mammalian cells to be members of a family of DNA hydroxylases that possess enzymatic activity toward the methyl mark on the 5-position of cytosine (5-methylcytosine [5mC]), a well-characterized epigenetic modification that has essential roles in regulating gene expression and maintaining cellular identity. Tet proteins can convert 5mC into 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) through three consecutive oxidation reactions. These modified bases may represent new epigenetic states in genomic DNA or intermediates in the process of DNA demethylation. Emerging biochemical, genetic, and functional evidence suggests that Tet proteins are crucial for diverse biological processes, including zygotic epigenetic reprogramming, pluripotent stem cell differentiation, hematopoiesis, and development of leukemia. Insights into how Tet proteins contribute to dynamic changes in DNA methylation and gene expression will greatly enhance our understanding of epigenetic regulation of normal development and human diseases.
Figures


References
-
- Abdel-Wahab O 2011. Genetics of the myeloproliferative neoplasms. Curr Opin Hematol 18: 117–123 - PubMed
-
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23: 185–188 - PubMed
-
- Bestor T, Laudano A, Mattaliano R, Ingram V 1988. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol 203: 971–983 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
