Structural activation of the transcriptional repressor EthR from Mycobacterium tuberculosis by single amino acid change mimicking natural and synthetic ligands
- PMID: 22156370
- PMCID: PMC3326297
- DOI: 10.1093/nar/gkr1113
Structural activation of the transcriptional repressor EthR from Mycobacterium tuberculosis by single amino acid change mimicking natural and synthetic ligands
Abstract
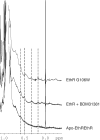
Ethionamide is an antituberculous drug for the treatment of multidrug-resistant Mycobacterium tuberculosis. This antibiotic requires activation by the monooxygenase EthA to exert its activity. Production of EthA is controlled by the transcriptional repressor EthR, a member of the TetR family. The sensitivity of M. tuberculosis to ethionamide can be artificially enhanced using synthetic ligands of EthR that allosterically inactivate its DNA-binding activity. Comparison of several structures of EthR co-crystallized with various ligands suggested that the structural reorganization of EthR resulting in its inactivation is controlled by a limited portion of the ligand-binding-pocket. In silico simulation predicted that mutation G106W may mimic ligands. X-ray crystallography of variant G106W indeed revealed a protein structurally similar to ligand-bound EthR. Surface plasmon resonance experiments established that this variant is unable to bind DNA, while thermal shift studies demonstrated that mutation G106W stabilizes EthR as strongly as ligands. Proton NMR of the methyl regions showed a lesser contribution of exchange broadening upon ligand binding, and the same quenched dynamics was observed in apo-variant G106W. Altogether, we here show that the area surrounding Gly106 constitutes the molecular switch involved in the conformational reorganization of EthR. These results also shed light on the mechanistic of ligand-induced allosterism controlling the DNA binding properties of TetR family repressors.
Figures

Similar articles
-
EthR, a repressor of the TetR/CamR family implicated in ethionamide resistance in mycobacteria, octamerizes cooperatively on its operator.Mol Microbiol. 2004 Jan;51(1):175-88. doi: 10.1046/j.1365-2958.2003.03809.x. Mol Microbiol. 2004. PMID: 14651620
-
Crystal structure of the TetR/CamR family repressor Mycobacterium tuberculosis EthR implicated in ethionamide resistance.J Mol Biol. 2004 Jul 23;340(5):1095-105. doi: 10.1016/j.jmb.2004.06.003. J Mol Biol. 2004. PMID: 15236969
-
A comprehensive analysis of the protein-ligand interactions in crystal structures of Mycobacterium tuberculosis EthR.Biochim Biophys Acta Proteins Proteom. 2019 Mar;1867(3):248-258. doi: 10.1016/j.bbapap.2018.12.003. Epub 2018 Dec 13. Biochim Biophys Acta Proteins Proteom. 2019. PMID: 30553830
-
Insights into mechanisms of induction and ligands recognition in the transcriptional repressor EthR from Mycobacterium tuberculosis.Tuberculosis (Edinb). 2006 Mar;86(2):110-4. doi: 10.1016/j.tube.2005.07.005. Epub 2005 Oct 21. Tuberculosis (Edinb). 2006. PMID: 16243584 Review.
-
Mycobacterium tuberculosis topoisomerases and EthR as the targets for new anti-TB drugs development.Future Med Chem. 2019 Aug;11(16):2193-2203. doi: 10.4155/fmc-2018-0232. Future Med Chem. 2019. PMID: 31538522 Review.
Cited by
-
Withdrawn.Infect Disord Drug Targets. 2012 Nov 16. Online ahead of print. Infect Disord Drug Targets. 2012. PMID: 23167715 Free PMC article.
-
Identification of a phenyl ester covalent inhibitor of caseinolytic protease and analysis of the ClpP1P2 inhibition in mycobacteria.mLife. 2025 Apr 15;4(2):155-168. doi: 10.1002/mlf2.12169. eCollection 2025 Apr. mLife. 2025. PMID: 40313980 Free PMC article.
-
Pre-Clinical Tools for Predicting Drug Efficacy in Treatment of Tuberculosis.Microorganisms. 2022 Feb 26;10(3):514. doi: 10.3390/microorganisms10030514. Microorganisms. 2022. PMID: 35336089 Free PMC article. Review.
-
Drug Resistance Mechanisms in Mycobacterium tuberculosis.Antibiotics (Basel). 2014 Jul 2;3(3):317-40. doi: 10.3390/antibiotics3030317. Antibiotics (Basel). 2014. PMID: 27025748 Free PMC article. Review.
-
Strategies for potentiation of ethionamide and folate antagonists against Mycobacterium tuberculosis.Expert Rev Anti Infect Ther. 2012 Sep;10(9):971-81. doi: 10.1586/eri.12.87. Expert Rev Anti Infect Ther. 2012. PMID: 23106273 Free PMC article. Review.
References
-
- Cox HS, Ford N, Reeder JC. Are we really that good at treating tuberculosis? Lancet Infect. Dis. 2009;9:138–139. - PubMed
-
- Frénois F, Engohang-Ndong J, Locht C, Baulard AR, Villeret V. Structure of EthR in a ligand bound conformation reveals therapeutic perspectives against tuberculosis. Mol. Cell. 2004;16:301–307. - PubMed
-
- Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. - PubMed
-
- Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 1996;2:662–667. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions

