A highly conserved residue in the C-terminal helix of HIV-1 matrix is required for envelope incorporation into virus particles
- PMID: 22156517
- PMCID: PMC3302379
- DOI: 10.1128/JVI.06047-11
A highly conserved residue in the C-terminal helix of HIV-1 matrix is required for envelope incorporation into virus particles
Abstract
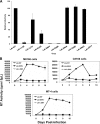
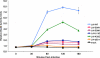
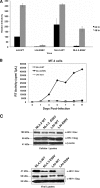
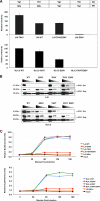
The incorporation of viral envelope (Env) glycoproteins into nascent particles is an essential step in the production of infectious human immunodeficiency virus type 1 (HIV-1). This process has been shown to require interactions between Env and the matrix (MA) domain of the Gag polyprotein. Previous studies indicate that several residues in the N-terminal region of MA are required for Env incorporation. However, the precise mechanism by which Env proteins are acquired during virus assembly has yet to be fully defined. Here, we examine whether a highly conserved glutamate at position 99 in the C-terminal helix is required for MA function and HIV-1 replication. We analyze a panel of mutant viruses that contain different amino acid substitutions at this position using viral infectivity studies, virus-cell fusion assays, and immunoblotting. We find that E99V mutant viruses are defective for fusion with cell membranes and thus are noninfectious. We show that E99V mutant particles of HIV-1 strains LAI and NL4.3 lack wild-type levels of Env proteins. We identify a compensatory substitution in MA residue 84 and show that it can reverse the E99V-associated defects. Taken together, these results indicate that the C-terminal hydrophobic pocket of MA, which encompasses both residues 84 and 99, has a previously unsuspected and key role in HIV-1 Env incorporation.
Figures




Similar articles
-
The role of matrix in HIV-1 envelope glycoprotein incorporation.Trends Microbiol. 2014 Jul;22(7):372-8. doi: 10.1016/j.tim.2014.04.012. Epub 2014 Jun 2. Trends Microbiol. 2014. PMID: 24933691 Free PMC article. Review.
-
HIV-1 Matrix Trimerization-Impaired Mutants Are Rescued by Matrix Substitutions That Enhance Envelope Glycoprotein Incorporation.J Virol. 2019 Dec 12;94(1):e01526-19. doi: 10.1128/JVI.01526-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31619553 Free PMC article.
-
Single-molecule imaging of HIV-1 envelope glycoprotein dynamics and Gag lattice association exposes determinants responsible for virus incorporation.Proc Natl Acad Sci U S A. 2019 Dec 10;116(50):25269-25277. doi: 10.1073/pnas.1910008116. Epub 2019 Nov 22. Proc Natl Acad Sci U S A. 2019. PMID: 31757854 Free PMC article.
-
Trimer Enhancement Mutation Effects on HIV-1 Matrix Protein Binding Activities.J Virol. 2016 May 27;90(12):5657-5664. doi: 10.1128/JVI.00509-16. Print 2016 Jun 15. J Virol. 2016. PMID: 27030269 Free PMC article.
-
The Interplay between HIV-1 Gag Binding to the Plasma Membrane and Env Incorporation.Viruses. 2020 May 16;12(5):548. doi: 10.3390/v12050548. Viruses. 2020. PMID: 32429351 Free PMC article. Review.
Cited by
-
Biological and biochemical characterization of HIV-1 Gag/dgp41 virus-like particles expressed in Nicotiana benthamiana.Plant Biotechnol J. 2013 Aug;11(6):681-90. doi: 10.1111/pbi.12058. Epub 2013 Mar 19. Plant Biotechnol J. 2013. PMID: 23506331 Free PMC article.
-
Clustering and mobility of HIV-1 Env at viral assembly sites predict its propensity to induce cell-cell fusion.J Virol. 2013 Jul;87(13):7516-25. doi: 10.1128/JVI.00790-13. Epub 2013 May 1. J Virol. 2013. PMID: 23637402 Free PMC article.
-
Role of the HIV-1 Matrix Protein in Gag Intracellular Trafficking and Targeting to the Plasma Membrane for Virus Assembly.Front Microbiol. 2012 Feb 17;3:55. doi: 10.3389/fmicb.2012.00055. eCollection 2012. Front Microbiol. 2012. PMID: 22363329 Free PMC article.
-
The role of matrix in HIV-1 envelope glycoprotein incorporation.Trends Microbiol. 2014 Jul;22(7):372-8. doi: 10.1016/j.tim.2014.04.012. Epub 2014 Jun 2. Trends Microbiol. 2014. PMID: 24933691 Free PMC article. Review.
-
Exosomes derived from HIV-1-infected cells contain trans-activation response element RNA.J Biol Chem. 2013 Jul 5;288(27):20014-33. doi: 10.1074/jbc.M112.438895. Epub 2013 May 9. J Biol Chem. 2013. PMID: 23661700 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

