High-mannose glycan-dependent epitopes are frequently targeted in broad neutralizing antibody responses during human immunodeficiency virus type 1 infection
- PMID: 22156525
- PMCID: PMC3302386
- DOI: 10.1128/JVI.06201-11
High-mannose glycan-dependent epitopes are frequently targeted in broad neutralizing antibody responses during human immunodeficiency virus type 1 infection
Abstract
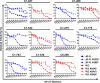
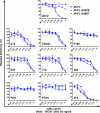
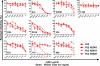
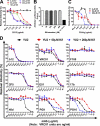
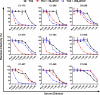
Broad and potent neutralizing antibody (BNAb) responses are rare in people infected by human immunodeficiency virus type 1 (HIV-1). Clearly defining the nature of BNAb epitopes on HIV-1 envelope glycoproteins (Envs) targeted in vivo is critical for future directions of anti-HIV-1 vaccine development. Conventional techniques are successful in defining neutralizing epitopes in a small number of individual subjects but fail in studying large groups of subjects. Two independent methods were employed to investigate the nature of NAb epitopes targeted in 9 subjects, identified by the NIAID Center for HIV/AIDS Vaccine Immunology (CHAVI) 001 and 008 clinical teams, known to make a strong BNAb response. Neutralizing activity from 8/9 subjects was enhanced by enriching high-mannose N-linked glycan (HM-glycan) of HIV-1 glycoproteins on neutralization target viruses and was sensitive to specific glycan deletion mutations of HIV-1 glycoproteins, indicating that HM-glycan-dependent epitopes are targeted by BNAb responses in these subjects. This discovery adds to accumulating evidence supporting the hypothesis that glycans are important targets on HIV-1 glycoproteins for BNAb responses in vivo, providing an important lead for future directions in developing NAb-based anti-HIV-1 vaccines.
Figures
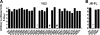
Similar articles
-
Vaccine Elicitation of High Mannose-Dependent Neutralizing Antibodies against the V3-Glycan Broadly Neutralizing Epitope in Nonhuman Primates.Cell Rep. 2017 Feb 28;18(9):2175-2188. doi: 10.1016/j.celrep.2017.02.003. Cell Rep. 2017. PMID: 28249163 Free PMC article.
-
Conformational Epitope-Specific Broadly Neutralizing Plasma Antibodies Obtained from an HIV-1 Clade C-Infected Elite Neutralizer Mediate Autologous Virus Escape through Mutations in the V1 Loop.J Virol. 2016 Jan 13;90(7):3446-57. doi: 10.1128/JVI.03090-15. J Virol. 2016. PMID: 26763999 Free PMC article.
-
Positive Selection at Key Residues in the HIV Envelope Distinguishes Broad and Strain-Specific Plasma Neutralizing Antibodies.J Virol. 2019 Mar 5;93(6):e01685-18. doi: 10.1128/JVI.01685-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30567996 Free PMC article.
-
Antibody responses to the HIV-1 envelope high mannose patch.Adv Immunol. 2019;143:11-73. doi: 10.1016/bs.ai.2019.08.002. Epub 2019 Sep 11. Adv Immunol. 2019. PMID: 31607367 Free PMC article. Review.
-
Beyond glycan barriers: non-cognate ligands and protein mimicry approaches to elicit broadly neutralizing antibodies for HIV-1.J Biomed Sci. 2024 Aug 21;31(1):83. doi: 10.1186/s12929-024-01073-y. J Biomed Sci. 2024. PMID: 39169357 Free PMC article. Review.
Cited by
-
A systematic study of the N-glycosylation sites of HIV-1 envelope protein on infectivity and antibody-mediated neutralization.Retrovirology. 2013 Feb 6;10:14. doi: 10.1186/1742-4690-10-14. Retrovirology. 2013. PMID: 23384254 Free PMC article.
-
Broadly Neutralizing Antibody Responses in a Large Longitudinal Sub-Saharan HIV Primary Infection Cohort.PLoS Pathog. 2016 Jan 14;12(1):e1005369. doi: 10.1371/journal.ppat.1005369. eCollection 2016 Jan. PLoS Pathog. 2016. PMID: 26766578 Free PMC article.
-
Algal lectins as potential HIV microbicide candidates.Mar Drugs. 2012 Jul;10(7):1476-1497. doi: 10.3390/md10071476. Epub 2012 Jul 10. Mar Drugs. 2012. PMID: 22851920 Free PMC article. Review.
-
Diversity and Function of Maternal HIV-1-Specific Antibodies at the Time of Vertical Transmission.J Virol. 2020 Apr 16;94(9):e01594-19. doi: 10.1128/JVI.01594-19. Print 2020 Apr 16. J Virol. 2020. PMID: 32075936 Free PMC article.
-
Glycan clustering stabilizes the mannose patch of HIV-1 and preserves vulnerability to broadly neutralizing antibodies.Nat Commun. 2015 Jun 24;6:7479. doi: 10.1038/ncomms8479. Nat Commun. 2015. PMID: 26105115 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

