The Andes hantavirus NSs protein is expressed from the viral small mRNA by a leaky scanning mechanism
- PMID: 22156529
- PMCID: PMC3302399
- DOI: 10.1128/JVI.06223-11
The Andes hantavirus NSs protein is expressed from the viral small mRNA by a leaky scanning mechanism
Abstract
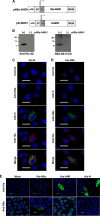
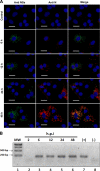
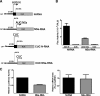
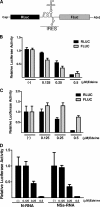
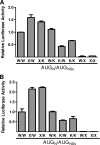
The small mRNA (SmRNA) of all Bunyaviridae encodes the nucleocapsid (N) protein. In 4 out of 5 genera in the Bunyaviridae, the smRNA encodes an additional nonstructural protein denominated NSs. In this study, we show that Andes hantavirus (ANDV) SmRNA encodes an NSs protein. Data show that the NSs protein is expressed in the context of an ANDV infection. Additionally, our results suggest that translation initiation from the NSs initiation codon is mediated by ribosomal subunits that have bypassed the upstream N protein initiation codon through a leaky scanning mechanism.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

