SILEC: a protocol for generating and using isotopically labeled coenzyme A mass spectrometry standards
- PMID: 22157971
- PMCID: PMC3802537
- DOI: 10.1038/nprot.2011.421
SILEC: a protocol for generating and using isotopically labeled coenzyme A mass spectrometry standards
Abstract
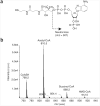
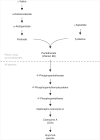
Stable isotope labeling by essential nutrients in cell culture (SILEC) was recently developed to generate isotopically labeled coenzyme A (CoA) and short-chain acyl-CoA thioesters. This was accomplished by modifying the widely used technique of stable isotope labeling by amino acids in cell culture to include [(13)C(3)(15)N]-pantothenate (vitamin B(5)), a CoA precursor, instead of the isotopically labeled amino acids. The lack of a de novo pantothenate synthesis pathway allowed for efficient and near-complete labeling of the measured CoA species. This protocol provides a step-by-step approach for generating stable isotope-labeled short-chain acyl-CoA internal standards in mammalian and insect cells as well as instructions on how to use them in stable isotope dilution mass spectrometric-based analyses. Troubleshooting guidelines, as well as a list of unlabeled and labeled CoA species, are also included. This protocol represents a prototype for generating stable isotope internal standards from labeled essential nutrients such as pantothenate. The generation and use of SILEC standards takes approximately 2-3 weeks.
Figures

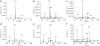

References
-
- Hsieh Y. HPLC-MS/MS in drug metabolism and pharmacokinetic screening. Expert Opin Drug Metab Toxicol. 2008;4:93–101. - PubMed
-
- Brown SC, Kruppa G, Dasseux JL. Metabolomics applications of FT-ICR mass spectrometry. Mass Spectrom Rev. 2005;24:223–231. - PubMed
-
- Hu QZ, et al. The Orbitrap: a new mass spectrometer. J Mass Spectrom. 2005;40:430–443. - PubMed
-
- Whalen K, Gobey J, Janiszewski J. A centralized approach to tandem mass spectrometry method development for high-throughput ADME screening. Rapid Commun Mass Spectrom. 2006;20:1497–1503. - PubMed
-
- Taylor CF, et al. A systematic approach to modeling, capturing, and disseminating proteomics experimental data. Nat Biotechnol. 2003;21:247–254. - PubMed
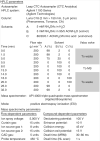
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

