Enhanced killing of cancer cells by poly(ADP-ribose) polymerase inhibitors and topoisomerase I inhibitors reflects poisoning of both enzymes
- PMID: 22158865
- PMCID: PMC3281688
- DOI: 10.1074/jbc.M111.296475
Enhanced killing of cancer cells by poly(ADP-ribose) polymerase inhibitors and topoisomerase I inhibitors reflects poisoning of both enzymes
Abstract
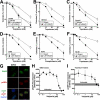
Poly(ADP-ribose) polymerase-1 (PARP1) plays critical roles in the regulation of DNA repair. Accordingly, small molecule inhibitors of PARP are being developed as agents that could modulate the activity of genotoxic chemotherapy, such as topoisomerase I poisons. In this study we evaluated the ability of the PARP inhibitor veliparib to enhance the cytotoxicity of the topoisomerase I poisons topotecan and camptothecin (CPT). Veliparib increased the cell cycle and cytotoxic effects of topotecan in multiple cell line models. Importantly, this sensitization occurred at veliparib concentrations far below those required to substantially inhibit poly(ADP-ribose) polymer synthesis and at least an order of magnitude lower than those involved in selective killing of homologous recombination-deficient cells. Further studies demonstrated that veliparib enhanced the effects of CPT in wild-type mouse embryonic fibroblasts (MEFs) but not Parp1(-/-) MEFs, confirming that PARP1 is the critical target for this sensitization. Importantly, parental and Parp1(-/-) MEFs had indistinguishable CPT sensitivities, ruling out models in which PARP1 catalytic activity plays a role in protecting cells from topoisomerase I poisons. To the contrary, cells were sensitized to CPT in a veliparib-independent manner upon transfection with PARP1 E988K, which lacks catalytic activity, or the isolated PARP1 DNA binding domain. These results are consistent with a model in which small molecule inhibitors convert PARP1 into a protein that potentiates the effects of topoisomerase I poisons by binding to damaged DNA and preventing its normal repair.
Figures



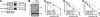

References
-
- Potmesil M., Hsiang Y. H., Liu L. F., Bank B., Grossberg H., Kirschenbaum S., Forlenza T. J., Penziner A., Kanganis D., Forlenzar T. J. (1988) Resistance of human leukemic and normal lymphocytes to drug-induced DNA cleavage and low levels of DNA topoisomerase II. Cancer Res. 48, 3537–3543 - PubMed
-
- Champoux J. J. (2001) DNA topoisomerases. Structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 - PubMed
-
- Muller M. T. (1985) Quantitation of eukaryotic topoisomerase I reactivity with DNA. Preferential cleavage of supercoiled DNA. Biochim. Biophys. Acta 824, 263–267 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

