Small molecule structure correctors abolish detrimental effects of apolipoprotein E4 in cultured neurons
- PMID: 22158868
- PMCID: PMC3285306
- DOI: 10.1074/jbc.M111.276162
Small molecule structure correctors abolish detrimental effects of apolipoprotein E4 in cultured neurons
Abstract
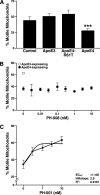
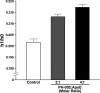
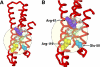
Apolipoprotein E4 (apoE4), the major genetic risk factor for late onset Alzheimer disease, assumes a pathological conformation, intramolecular domain interaction. ApoE4 domain interaction mediates the detrimental effects of apoE4, including decreased mitochondrial cytochrome c oxidase subunit 1 levels, reduced mitochondrial motility, and reduced neurite outgrowth in vitro. Mutant apoE4 (apoE4-R61T) lacks domain interaction, behaves like apoE3, and does not cause detrimental effects. To identify small molecules that inhibit domain interaction (i.e. structure correctors) and reverse the apoE4 detrimental effects, we established a high throughput cell-based FRET primary assay that determines apoE4 domain interaction and secondary cell- and function-based assays. Screening a ChemBridge library with the FRET assay identified CB9032258 (a phthalazinone derivative), which inhibits domain interaction in neuronal cells. In secondary functional assays, CB9032258 restored mitochondrial cytochrome c oxidase subunit 1 levels and rescued impairments of mitochondrial motility and neurite outgrowth in apoE4-expressing neuronal cells. These benefits were apoE4-specific and dose-dependent. Modifying CB9032258 yielded well defined structure-activity relationships and more active compounds with enhanced potencies in the FRET assay (IC(50) of 23 and 116 nm, respectively). These compounds efficiently restored functional activities of apoE4-expressing cells in secondary assays. An EPR binding assay showed that the apoE4 structure correction resulted from direct interaction of a phthalazinone. With these data, a six-feature pharmacophore model was constructed for future drug design. Our results serve as a proof of concept that pharmacological intervention with apoE4 structure correctors negates apoE4 detrimental effects in neuronal cells and could be further developed as an Alzheimer disease therapeutic.
Figures






References
-
- Corder E. H., Saunders A. M., Strittmatter W. J., Schmechel D. E., Gaskell P. C., Small G. W., Roses A. D., Haines J. L., Pericak-Vance M. A. (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261, 921–923 - PubMed
-
- Strittmatter W. J., Saunders A. M., Schmechel D., Pericak-Vance M., Enghild J., Salvesen G. S., Roses A. D. (1993) Apolipoprotein E. High-avidity binding to β-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 90, 1977–1981 - PMC - PubMed
-
- Saunders A. M., Strittmatter W. J., Schmechel D., St George-Hyslop P. H., Pericak-Vance M. A., Joo S. H., Rosi B. L., Gusella J. F., Crapper-MacLachlan D. R., Alberts M. J., Hulette C., Crain B., Goldgaber D., Roses A. D. (1993) Association of apolipoprotein E allele ϵ4 with late-onset familial and sporadic Alzheimer's disease. Neurology 43, 1467–1472 - PubMed
-
- Farrer L. A., Cupples L. A., Haines J. L., Hyman B., Kukull W. A., Mayeux R., Myers R. H., Pericak-Vance M. A., Risch N., van Duijn C. M. (1997) Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. JAMA 278, 1349–1356 - PubMed
-
- Mahley R. W. (1988) Apolipoprotein E. Cholesterol transport protein with expanding role in cell biology. Science 240, 622–630 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

