Niche specialization of terrestrial archaeal ammonia oxidizers
- PMID: 22158986
- PMCID: PMC3248517
- DOI: 10.1073/pnas.1109000108
Niche specialization of terrestrial archaeal ammonia oxidizers
Abstract
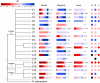
Soil pH is a major determinant of microbial ecosystem processes and potentially a major driver of evolution, adaptation, and diversity of ammonia oxidizers, which control soil nitrification. Archaea are major components of soil microbial communities and contribute significantly to ammonia oxidation in some soils. To determine whether pH drives evolutionary adaptation and community structure of soil archaeal ammonia oxidizers, sequences of amoA, a key functional gene of ammonia oxidation, were examined in soils at global, regional, and local scales. Globally distributed database sequences clustered into 18 well-supported phylogenetic lineages that dominated specific soil pH ranges classified as acidic (pH <5), acido-neutral (5 ≤ pH <7), or alkalinophilic (pH ≥ 7). To determine whether patterns were reproduced at regional and local scales, amoA gene fragments were amplified from DNA extracted from 47 soils in the United Kingdom (pH 3.5-8.7), including a pH-gradient formed by seven soils at a single site (pH 4.5-7.5). High-throughput sequencing and analysis of amoA gene fragments identified an additional, previously undiscovered phylogenetic lineage and revealed similar pH-associated distribution patterns at global, regional, and local scales, which were most evident for the five most abundant clusters. Archaeal amoA abundance and diversity increased with soil pH, which was the only physicochemical characteristic measured that significantly influenced community structure. These results suggest evolution based on specific adaptations to soil pH and niche specialization, resulting in a global distribution of archaeal lineages that have important consequences for soil ecosystem function and nitrogen cycling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high toxicity in soil. Science. 2005;309:1387–1390. - PubMed
-
- Philippot L, et al. The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol. 2010;8:523–529. - PubMed
-
- Griffiths RI, et al. The bacterial biogeography of British soils. Environ Microbiol. 2011;13:1642–1654. - PubMed
-
- Valentine DL. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat Rev Microbiol. 2007;5:316–323. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

