Functional characterization of glycine N-methyltransferase and its interactive protein DEPDC6/DEPTOR in hepatocellular carcinoma
- PMID: 22160218
- PMCID: PMC3324955
- DOI: 10.2119/molmed.2011.00331
Functional characterization of glycine N-methyltransferase and its interactive protein DEPDC6/DEPTOR in hepatocellular carcinoma
Erratum in
- Mol Med. 2012;18(1):546
Abstract
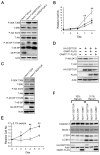
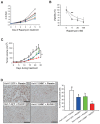
Glycine N-methyltransferase (GNMT) is a tumor suppressor for hepatocellular carcinoma (HCC). High rates of Gnmt knockout mice developed HCC. Epigenetic alteration and dysregulation of several pathways including wingless-type MMTV integration site (Wnt), mitogen-activated protein kinase (MAPK) and Janus kinase and signal transducer and activator of transcription (JAK-STAT) are associated with HCC development in Gnmt knockout mice. We hypothesized that GNMT may regulate signal transduction through interacting with other proteins directly. In this report, we identified a mammalian target of rapamycin (mTOR) inhibitor (DEP domain containing MTOR-interacting protein [DEPDC6/DEPTOR]) as a GNMT-binding protein by using yeast two-hybrid screening. Fluorescence resonance energy transfer assay demonstrated that the C-terminal half of GNMT interact with the PSD-95/Dlg1/ZO-1 (PDZ) domain of DEPDC6/DEPTOR. Immunohistochemical staining showed that 27.5% (14/51) of HCC patients had higher expression levels of DEPDC6/DEPTOR in the tumorous tissues than in tumor-adjacent tissues, especially among HCC patients with hepatitis B viral infection (odds ratio 10.3, 95% confidence interval [CI] 1.05-11.3) or patients with poor prognosis (death hazard ratio 4.51, 95% CI 1.60-12.7). In terms of molecular mechanism, knockdown of DEPDC6/DEPTOR expression in HuH-7 cells caused S6K and 4E-BP activation, but suppressed Akt. Overexpression of DEPDC6/DEPTOR activated Akt and increased survival of HCC cells. Overexpression of GNMT caused activation of mTOR/raptor downstream signaling and delayed G2/M cell cycle progression, which altogether resulted in cellular senescence. Furthermore, GNMT reduced proliferation of HuH-7 cells and sensitized them to rapamycin treatment both in vitro and in vivo. In conclusion, GNMT regulates HCC growth in part through interacting with DEPDC6/DEPTOR and modulating mTOR/raptor signaling pathway. Both GNMT and DEPDC6/DEPTOR are potential targets for developing therapeutics for HCC.
Figures
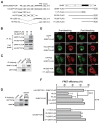


 , HuH-7-GFP + 4;
, HuH-7-GFP + 4;
 , HuH-7-GFP + 20;
, HuH-7-GFP + 20;
 , HuH-7-GFP + 100; ●, HuH-7-GNMT + 0;
, HuH-7-GFP + 100; ●, HuH-7-GNMT + 0;
 , HuH-7-GNMT + 4;
, HuH-7-GNMT + 4;
 , HuH-7-GNMT + 20;
, HuH-7-GNMT + 20;
 , HuH-7-GNMT + 100; panel B: —○—, HuH-7-GFP; ---●---, HuH-7-GNMT; panel C: —○—, HuH-7-GFP + placebo;
, HuH-7-GNMT + 100; panel B: —○—, HuH-7-GFP; ---●---, HuH-7-GNMT; panel C: —○—, HuH-7-GFP + placebo;
 , HuH-7-GFP + RAD001;
, HuH-7-GFP + RAD001;
 , HuH-7-GNMT + placebo;
, HuH-7-GNMT + placebo;
 , HuH-7-GNMT + RAD001.
, HuH-7-GNMT + RAD001.References
-
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. - PubMed
-
- Finn RS. Development of molecularly targeted therapies in hepatocellular carcinoma: where do we go now? Clin Cancer Res. 2010;16:390–7. - PubMed
-
- Sahin F, et al. mTOR and P70 S6 kinase expression in primary liver neoplasms. Clin Cancer Res. 2004;10:8421–5. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

