Yeast formin Bni1p has multiple localization regions that function in polarized growth and spindle orientation
- PMID: 22160598
- PMCID: PMC3268721
- DOI: 10.1091/mbc.E11-07-0631
Yeast formin Bni1p has multiple localization regions that function in polarized growth and spindle orientation
Abstract
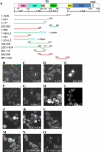
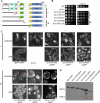
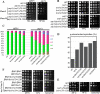
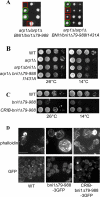
Formins are conserved proteins that assemble unbranched actin filaments in a regulated, localized manner. Budding yeast's two formins, Bni1p and Bnr1p, assemble actin cables necessary for polarized cell growth and organelle segregation. Here we define four regions in Bni1p that contribute to its localization to the bud and at the bud neck. The first (residues 1-333) requires dimerization for its localization and encompasses the Rho-binding domain. The second (residues 334-821) covers the Diaphanous inhibitory-dimerization-coiled coil domains, and the third is the Spa2p-binding domain. The fourth region encompasses the formin homology 1-formin homology 2-COOH region of the protein. These four regions can each localize to the bud cortex and bud neck at the right stage of the cell cycle independent of both F-actin and endogenous Bni1p. The first three regions contribute cumulatively to the proper localization of Bni1p, as revealed by the effects of progressive loss of these regions on the actin cytoskeleton and fidelity of spindle orientation. The fourth region contributes to the localization of Bni1p in tiny budded cells. Expression of mislocalized Bni1p constructs has a dominant-negative effect on both growth and nuclear segregation due to mislocalized actin assembly. These results define an unexpected complexity in the mechanism of formin localization and function.
Figures

References
-
- Beach DL, Thibodeaux J, Maddox P, Yeh E, Bloom K. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr Biol. 2000;10:1497–1506. - PubMed
-
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. - PubMed
-
- Burbelo PD, Drechsel D, Hall A. A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J Biol Chem. 1995;270:29071–29074. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

