Variable motif utilization in homeotic selector (Hox)-cofactor complex formation controls specificity
- PMID: 22160705
- PMCID: PMC3248519
- DOI: 10.1073/pnas.1114118109
Variable motif utilization in homeotic selector (Hox)-cofactor complex formation controls specificity
Abstract
Homeotic selector (Hox) proteins often bind DNA cooperatively with cofactors such as Extradenticle (Exd) and Homothorax (Hth) to achieve functional specificity in vivo. Previous studies identified the Hox YPWM motif as an important Exd interaction motif. Using a comparative approach, we characterize the contribution of this and additional conserved sequence motifs to the regulation of specific target genes for three Drosophila Hox proteins. We find that Sex combs reduced (Scr) uses a simple interaction mechanism, where a single tryptophan-containing motif is necessary for Exd-dependent DNA-binding and in vivo functions. Abdominal-A (AbdA) is more complex, using multiple conserved motifs in a context-dependent manner. Lastly, Ultrabithorax (Ubx) is the most flexible, in that it uses multiple conserved motifs that function in parallel to regulate target genes in vivo. We propose that using different binding mechanisms with the same cofactor may be one strategy to achieve functional specificity in vivo.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
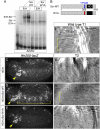
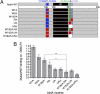
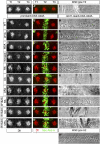
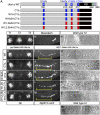
Similar articles
-
Functional synthetic Antennapedia genes and the dual roles of YPWM motif and linker size in transcriptional activation and repression.Proc Natl Acad Sci U S A. 2011 Jul 19;108(29):11959-64. doi: 10.1073/pnas.1108686108. Epub 2011 Jun 28. Proc Natl Acad Sci U S A. 2011. PMID: 21712439 Free PMC article.
-
Hox repression of a target gene: extradenticle-independent, additive action through multiple monomer binding sites.Development. 2002 Jul;129(13):3115-26. doi: 10.1242/dev.129.13.3115. Development. 2002. PMID: 12070087
-
Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle complex.Development. 1999 Nov;126(22):5137-48. doi: 10.1242/dev.126.22.5137. Development. 1999. PMID: 10529430
-
The specificity of homeotic gene function.Bioessays. 1995 Oct;17(10):855-63. doi: 10.1002/bies.950171007. Bioessays. 1995. PMID: 7487967 Review.
-
Hox cofactors in vertebrate development.Dev Biol. 2006 Mar 15;291(2):193-206. doi: 10.1016/j.ydbio.2005.10.032. Epub 2006 Mar 3. Dev Biol. 2006. PMID: 16515781 Review.
Cited by
-
Phenotypic Nonspecificity as the Result of Limited Specificity of Transcription Factor Function.Genet Res Int. 2018 Oct 28;2018:7089109. doi: 10.1155/2018/7089109. eCollection 2018. Genet Res Int. 2018. PMID: 30510805 Free PMC article. Review.
-
New Insights into Cooperative Binding of Homeodomain Transcription Factors PREP1 and PBX1 to DNA.Sci Rep. 2017 Jan 17;7:40665. doi: 10.1038/srep40665. Sci Rep. 2017. PMID: 28094776 Free PMC article.
-
Mechanisms of Specificity for Hox Factor Activity.J Dev Biol. 2016 Jun;4(2):16. doi: 10.3390/jdb4020016. Epub 2016 May 9. J Dev Biol. 2016. PMID: 27583210 Free PMC article.
-
Nuclear microenvironments modulate transcription from low-affinity enhancers.Elife. 2017 Nov 2;6:e28975. doi: 10.7554/eLife.28975. Elife. 2017. PMID: 29095143 Free PMC article.
-
Role of HOXA9 in leukemia: dysregulation, cofactors and essential targets.Oncogene. 2016 Mar 3;35(9):1090-8. doi: 10.1038/onc.2015.174. Epub 2015 Jun 1. Oncogene. 2016. PMID: 26028034 Free PMC article. Review.
References
-
- Gebelein B, McKay DJ, Mann RS. Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature. 2004;431:653–659. - PubMed
-
- Georges AB, Benayoun BA, Caburet S, Veitia RA. Generic binding sites, generic DNA-binding domains: Where does specific promoter recognition come from? FASEB J. 2010;24:346–356. - PubMed
-
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

