microRNA-34a regulates neurite outgrowth, spinal morphology, and function
- PMID: 22160706
- PMCID: PMC3248521
- DOI: 10.1073/pnas.1112063108
microRNA-34a regulates neurite outgrowth, spinal morphology, and function
Abstract
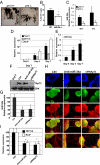
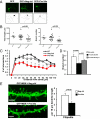
The p53 family member TAp73 is a transcription factor that plays a key role in many biological processes, including neuronal development. In particular, we have shown that p73 drives the expression of miR-34a, but not miR-34b and c, in mouse cortical neurons. miR-34a in turn modulates the expression of synaptic targets including synaptotagmin-1 and syntaxin-1A. Here we show that this axis is retained in mouse ES cells committed to differentiate toward a neurological phenotype. Moreover, overexpression of miR-34a alters hippocampal spinal morphology, and results in electrophysiological changes consistent with a reduction in spinal function. Therefore, the TAp73/miR-34a axis has functional relevance in primary neurons. These data reinforce a role for miR-34a in neuronal development.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Stefani G, Slack F-J. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. - PubMed
-
- Kosik K-S. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. - PubMed
-
- Kaghad M, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

