Epidermal growth factor receptor downregulation by small heterodimeric binding proteins
- PMID: 22160867
- PMCID: PMC3286197
- DOI: 10.1093/protein/gzr056
Epidermal growth factor receptor downregulation by small heterodimeric binding proteins
Abstract
No single engineered protein has been shown previously to robustly downregulate epidermal growth factor receptor (EGFR), a validated cancer target. A panel of fibronectin-based domains was engineered to bind with picomolar to nanomolar affinity to multiple epitopes of EGFR. Monovalent and homo- and hetero-bivalent dimers of these domains were tested for EGFR downregulation. Selected orientations of non-competitive heterodimers decrease EGFR levels by up to 80% in multiple cell types, without activating receptor signaling. These heterodimers inhibit autophosphorylation, proliferation and migration, and are synergistic with the monoclonal antibody cetuximab in these activities. These small (25 kDa) heterodimers represent a novel modality for modulating surface receptor levels.
Figures
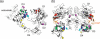
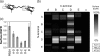



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

