Morphological defects in a novel Rdh10 mutant that has reduced retinoic acid biosynthesis and signaling
- PMID: 22162152
- PMCID: PMC4118640
- DOI: 10.1002/dvg.22002
Morphological defects in a novel Rdh10 mutant that has reduced retinoic acid biosynthesis and signaling
Abstract
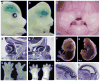
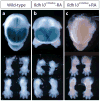
Retinoic acid (RA) signaling is necessary for proper patterning and morphogenesis during embryonic development. Tissue-specific RA signaling requires precise spatial and temporal synthesis of RA from retinal by retinaldehyde dehydrogenases (Raldh) and the conversion of retinol to retinal by retinol dehydrogenases (Rdh) of the short-chain dehydrogenase/reducatase gene family (SDR). The SDR, retinol dehydrogenase 10 (RDH10), is a major contributor to retinal biosynthesis during mid-gestation. We have identified a missense mutation in the Rdh10 gene (Rdh10(m366Asp) ) using an N-ethyl-N-nitrosourea-induced forward genetic screen that result in reduced RA levels and signaling during embryonic development. Rdh10(m366Asp) mutant embryos have unique phenotypes, such as edema, a massive midline facial cleft, and neurogenesis defects in the forebrain, that will allow the identification of novel RA functions.
Copyright © 2011 Wiley Periodicals, Inc.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

