Rapid identification of mycobacteria and drug-resistant Mycobacterium tuberculosis by use of a single multiplex PCR and DNA sequencing
- PMID: 22162548
- PMCID: PMC3264146
- DOI: 10.1128/JCM.05570-11
Rapid identification of mycobacteria and drug-resistant Mycobacterium tuberculosis by use of a single multiplex PCR and DNA sequencing
Abstract
Tuberculosis (TB) remains a significant global health problem for which rapid diagnosis is critical to both treatment and control. This report describes a multiplex PCR method, the Mycobacterial IDentification and Drug Resistance Screen (MID-DRS) assay, which allows identification of members of the Mycobacterium tuberculosis complex (MTBC) and the simultaneous amplification of targets for sequencing-based drug resistance screening of rifampin-resistant (rifampin(r)), isoniazid(r), and pyrazinamide(r) TB. Additionally, the same multiplex reaction amplifies a specific 16S rRNA gene target for rapid identification of M. avium complex (MAC) and a region of the heat shock protein 65 gene (hsp65) for further DNA sequencing-based confirmation or identification of other mycobacterial species. Comparison of preliminary results generated with MID-DRS versus culture-based methods for a total of 188 bacterial isolates demonstrated MID-DRS sensitivity and specificity as 100% and 96.8% for MTBC identification; 100% and 98.3% for MAC identification; 97.4% and 98.7% for rifampin(r) TB identification; 60.6% and 100% for isoniazid(r) TB identification; and 75.0% and 98.1% for pyrazinamide(r) TB identification. The performance of the MID-DRS was also tested on acid-fast-bacterium (AFB)-positive clinical specimens, resulting in sensitivity and specificity of 100% and 78.6% for detection of MTBC and 100% and 97.8% for detection of MAC. In conclusion, use of the MID-DRS reduces the time necessary for initial identification and drug resistance screening of TB specimens to as little as 2 days. Since all targets needed for completing the assay are included in a single PCR amplification step, assay costs, preparation time, and risks due to user errors are also reduced.
Figures
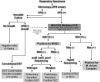


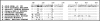
Similar articles
-
A Rapid Drug Resistance Genotyping Workflow for Mycobacterium tuberculosis, Using Targeted Isothermal Amplification and Nanopore Sequencing.Microbiol Spectr. 2021 Dec 22;9(3):e0061021. doi: 10.1128/Spectrum.00610-21. Epub 2021 Nov 24. Microbiol Spectr. 2021. PMID: 34817282 Free PMC article.
-
Evaluation of efficiency of nested multiplex allele-specific PCR assay for detection of multidrug resistant tuberculosis directly from sputum samples.Lett Appl Microbiol. 2016 May;62(5):411-8. doi: 10.1111/lam.12564. Lett Appl Microbiol. 2016. PMID: 26972777
-
Detection of Rifampicin- and Isoniazid-Resistant Mycobacterium tuberculosis Using the Quantamatrix Multiplexed Assay Platform System.Ann Lab Med. 2018 Nov;38(6):569-577. doi: 10.3343/alm.2018.38.6.569. Ann Lab Med. 2018. PMID: 30027701 Free PMC article.
-
Rapid diagnostics of tuberculosis and drug resistance in the industrialized world: clinical and public health benefits and barriers to implementation.BMC Med. 2013 Aug 29;11:190. doi: 10.1186/1741-7015-11-190. BMC Med. 2013. PMID: 23987891 Free PMC article. Review.
-
Multidrug-resistant tuberculosis drug susceptibility and molecular diagnostic testing.Am J Med Sci. 2013 Feb;345(2):143-8. doi: 10.1097/MAJ.0b013e31825d32c6. Am J Med Sci. 2013. PMID: 22885627 Free PMC article. Review.
Cited by
-
Rapid detection of Mycobacterium tuberculosis by recombinase polymerase amplification.PLoS One. 2014 Aug 13;9(8):e103091. doi: 10.1371/journal.pone.0103091. eCollection 2014. PLoS One. 2014. PMID: 25118698 Free PMC article.
-
Comparative Study of GeneXpert MTB/RIF Assay and Multiplex PCR Assay for Direct Detection of Mycobacterium tuberculosis in Suspected Pulmonary Tuberculosis Patients.Curr Microbiol. 2017 Sep;74(9):1026-1032. doi: 10.1007/s00284-017-1279-x. Epub 2017 Jun 13. Curr Microbiol. 2017. PMID: 28612135
-
Molecular surveillance of tuberculosis-causing mycobacteria in wastewater.Heliyon. 2022 Feb 4;8(2):e08910. doi: 10.1016/j.heliyon.2022.e08910. eCollection 2022 Feb. Heliyon. 2022. PMID: 35198775 Free PMC article.
-
Performance and workflow assessment of six nucleic acid extraction technologies for use in resource limited settings.PLoS One. 2019 Apr 18;14(4):e0215753. doi: 10.1371/journal.pone.0215753. eCollection 2019. PLoS One. 2019. PMID: 30998749 Free PMC article.
-
Diagnosis of osteoarticular tuberculosis via metagenomic next-generation sequencing: A case report.Exp Ther Med. 2019 Aug;18(2):1184-1188. doi: 10.3892/etm.2019.7655. Epub 2019 Jun 10. Exp Ther Med. 2019. PMID: 31316612 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

