Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl(-/-) retinal transcriptomes
- PMID: 22162623
- PMCID: PMC3233386
Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl(-/-) retinal transcriptomes
Abstract
Purpose: Next-generation sequencing (NGS) has revolutionized systems-based analysis of cellular pathways. The goals of this study are to compare NGS-derived retinal transcriptome profiling (RNA-seq) to microarray and quantitative reverse transcription polymerase chain reaction (qRT-PCR) methods and to evaluate protocols for optimal high-throughput data analysis.
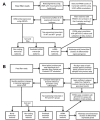
Methods: Retinal mRNA profiles of 21-day-old wild-type (WT) and neural retina leucine zipper knockout (Nrl(-/-)) mice were generated by deep sequencing, in triplicate, using Illumina GAIIx. The sequence reads that passed quality filters were analyzed at the transcript isoform level with two methods: Burrows-Wheeler Aligner (BWA) followed by ANOVA (ANOVA) and TopHat followed by Cufflinks. qRT-PCR validation was performed using TaqMan and SYBR Green assays.
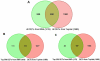
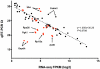
Results: Using an optimized data analysis workflow, we mapped about 30 million sequence reads per sample to the mouse genome (build mm9) and identified 16,014 transcripts in the retinas of WT and Nrl(-/-) mice with BWA workflow and 34,115 transcripts with TopHat workflow. RNA-seq data confirmed stable expression of 25 known housekeeping genes, and 12 of these were validated with qRT-PCR. RNA-seq data had a linear relationship with qRT-PCR for more than four orders of magnitude and a goodness of fit (R(2)) of 0.8798. Approximately 10% of the transcripts showed differential expression between the WT and Nrl(-/-) retina, with a fold change ≥1.5 and p value <0.05. Altered expression of 25 genes was confirmed with qRT-PCR, demonstrating the high degree of sensitivity of the RNA-seq method. Hierarchical clustering of differentially expressed genes uncovered several as yet uncharacterized genes that may contribute to retinal function. Data analysis with BWA and TopHat workflows revealed a significant overlap yet provided complementary insights in transcriptome profiling.
Conclusions: Our study represents the first detailed analysis of retinal transcriptomes, with biologic replicates, generated by RNA-seq technology. The optimized data analysis workflows reported here should provide a framework for comparative investigations of expression profiles. Our results show that NGS offers a comprehensive and more accurate quantitative and qualitative evaluation of mRNA content within a cell or tissue. We conclude that RNA-seq based transcriptome characterization would expedite genetic network analyses and permit the dissection of complex biologic functions.
Figures
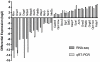

References
-
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. - PMC - PubMed
-
- Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–32. - PubMed
-
- Schuster SC. Next-generation sequencing transforms today's biology. Nat Methods. 2008;5:16–8. - PubMed
-
- Roukos DH. Next-generation sequencing and epigenome technologies: potential medical applications. Expert Rev Med Devices. 2010;7:723–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
