A human monoclonal antibody with neutralizing activity against highly divergent influenza subtypes
- PMID: 22162996
- PMCID: PMC3230632
- DOI: 10.1371/journal.pone.0028001
A human monoclonal antibody with neutralizing activity against highly divergent influenza subtypes
Abstract
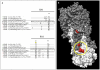
The interest in broad-range anti-influenza A monoclonal antibodies (mAbs) has recently been strengthened by the identification of anti-hemagglutinin (HA) mAbs endowed with heterosubtypic neutralizing activity to be used in the design of "universal" prophylactic or therapeutic tools. However, the majority of the single mAbs described to date do not bind and neutralize viral isolates belonging to highly divergent subtypes clustering into the two different HA-based influenza phylogenetic groups: the group 1 including, among others, subtypes H1, H2, H5 and H9 and the group 2 including, among others, H3 subtype. Here, we describe a human mAb, named PN-SIA28, capable of binding and neutralizing all tested isolates belonging to phylogenetic group 1, including H1N1, H2N2, H5N1 and H9N2 subtypes and several isolates belonging to group 2, including H3N2 isolates from the first period of the 1968 pandemic. Therefore, PN-SIA28 is capable of neutralizing isolates belonging to subtypes responsible of all the reported pandemics, as well as other subtypes with pandemic potential. The region recognized by PN-SIA28 has been identified on the stem region of HA and includes residues highly conserved among the different influenza subtypes. A deep characterization of PN-SIA28 features may represent a useful help in the improvement of available anti-influenza therapeutic strategies and can provide new tools for the development of universal vaccinal strategies.
Conflict of interest statement
Figures



References
-
- Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine. 2007;25:5086–5096. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

