Modulation of thyroid hormone-dependent gene expression in Xenopus laevis by INhibitor of Growth (ING) proteins
- PMID: 22163049
- PMCID: PMC3230625
- DOI: 10.1371/journal.pone.0028658
Modulation of thyroid hormone-dependent gene expression in Xenopus laevis by INhibitor of Growth (ING) proteins
Abstract
Background: INhibitor of Growth (ING) proteins belong to a large family of plant homeodomain finger-containing proteins important in epigenetic regulation and carcinogenesis. We have previously shown that ING1 and ING2 expression is regulated by thyroid hormone (TH) during metamorphosis of the Xenopus laevis tadpole. The present study investigates the possibility that ING proteins modulate TH action.
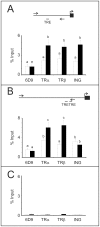
Methodology/principal findings: Tadpoles expressing a Xenopus ING2 transgene (Trans(ING2)) were significantly smaller than tadpoles not expressing the transgene (Trans(GFP)). When exposed to 10 nM 3,5,3'-triiodothyronine (T(3)), premetamorphic Trans(ING2) tadpoles exhibited a greater reduction in tail, head, and brain areas, and a protrusion of the lower jaw than T(3)-treated Trans(GFP) tadpoles. Quantitative real time polymerase chain reaction (QPCR) demonstrated elevated TH receptor β (TRβ) and TH/bZIP transcript levels in Trans(ING2) tadpole tails compared to Trans(GFP) tadpoles while TRα mRNAs were unaffected. In contrast, no difference in TRα, TRβ or insulin-like growth factor (IGF2) mRNA abundance was observed in the brain between Trans(ING2) and Trans(GFP) tadpoles. All of these transcripts, except for TRα mRNA in the brain, were inducible by the hormone in both tissues. Oocyte transcription assays indicated that ING proteins enhanced TR-dependent, T(3)-induced TRβ gene promoter activity. Examination of endogenous T(3)-responsive promoters (TRβ and TH/bZIP) in the tail by chromatin immunoprecipitation assays showed that ING proteins were recruited to TRE-containing regions in T(3)-dependent and independent ways, respectively. Moreover, ING and TR proteins coimmunoprecipitated from tail protein homogenates derived from metamorphic climax animals.
Conclusions/significance: We show for the first time that ING proteins modulate TH-dependent responses, thus revealing a novel role for ING proteins in hormone signaling. This has important implications for understanding hormone influenced disease states and suggests that the induction of ING proteins may facilitate TR function during metamorphosis in a tissue-specific manner.
Conflict of interest statement
Figures





Similar articles
-
Multiple ING1 and ING2 genes in Xenopus laevis and evidence for differential association of thyroid hormone receptors and ING proteins to their promoters.Biochim Biophys Acta. 2008 Mar;1779(3):152-63. doi: 10.1016/j.bbagrm.2007.12.002. Epub 2007 Dec 14. Biochim Biophys Acta. 2008. PMID: 18167318
-
Expression of novel ING variants is regulated by thyroid hormone in the Xenopus laevis tadpole.J Biol Chem. 2001 Dec 14;276(50):47013-20. doi: 10.1074/jbc.M106965200. Epub 2001 Oct 12. J Biol Chem. 2001. PMID: 11600495
-
Gene-specific changes in promoter occupancy by thyroid hormone receptor during frog metamorphosis. Implications for developmental gene regulation.J Biol Chem. 2005 Dec 16;280(50):41222-8. doi: 10.1074/jbc.M509593200. Epub 2005 Oct 19. J Biol Chem. 2005. PMID: 16236718
-
Autoinduction of nuclear receptor genes and its significance.J Steroid Biochem Mol Biol. 1993 Aug;46(2):105-19. doi: 10.1016/0960-0760(93)90286-6. J Steroid Biochem Mol Biol. 1993. PMID: 8664159 Review.
-
ING1 and ING2: multifaceted tumor suppressor genes.Cell Mol Life Sci. 2013 Oct;70(20):3753-72. doi: 10.1007/s00018-013-1270-z. Epub 2013 Feb 15. Cell Mol Life Sci. 2013. PMID: 23412501 Free PMC article. Review.
References
-
- Menendez C, Abad M, Gomez-Cabello D, Moreno A, Palmero I. ING proteins in cellular senescence. Curr Drug Targets. 2009;10:406–417. - PubMed
-
- Li J, Wang Y, Wong RP, Li G. The role of ING tumor suppressors in UV stress response and melanoma progression. Curr Drug Targets. 2009;10:455–464. - PubMed
-
- Gunduz M, Demircan K, Gunduz E, Katase N, Tamamura R, Nagatsuka H. Potential usage of ING family members in cancer diagnostics and molecular therapy. Curr Drug Targets. 2009;10:465–476. - PubMed
-
- Walzak AA, Veldhoen N, Feng X, Riabowol K, Helbing CC. Expression profiles of mRNA transcript variants encoding the human inhibitor of growth tumor suppressor gene family in normal and neoplastic tissues. Exp Cell Res. 2008;314:273–285. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

