Correlation network analysis applied to complex biofilm communities
- PMID: 22163302
- PMCID: PMC3233593
- DOI: 10.1371/journal.pone.0028438
Correlation network analysis applied to complex biofilm communities
Abstract
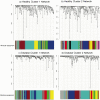
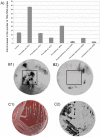
The complexity of the human microbiome makes it difficult to reveal organizational principles of the community and even more challenging to generate testable hypotheses. It has been suggested that in the gut microbiome species such as Bacteroides thetaiotaomicron are keystone in maintaining the stability and functional adaptability of the microbial community. In this study, we investigate the interspecies associations in a complex microbial biofilm applying systems biology principles. Using correlation network analysis we identified bacterial modules that represent important microbial associations within the oral community. We used dental plaque as a model community because of its high diversity and the well known species-species interactions that are common in the oral biofilm. We analyzed samples from healthy individuals as well as from patients with periodontitis, a polymicrobial disease. Using results obtained by checkerboard hybridization on cultivable bacteria we identified modules that correlated well with microbial complexes previously described. Furthermore, we extended our analysis using the Human Oral Microbe Identification Microarray (HOMIM), which includes a large number of bacterial species, among them uncultivated organisms present in the mouth. Two distinct microbial communities appeared in healthy individuals while there was one major type in disease. Bacterial modules in all communities did not overlap, indicating that bacteria were able to effectively re-associate with new partners depending on the environmental conditions. We then identified hubs that could act as keystone species in the bacterial modules. Based on those results we then cultured a not-yet-cultivated microorganism, Tannerella sp. OT286 (clone BU063). After two rounds of enrichment by a selected helper (Prevotella oris OT311) we obtained colonies of Tannerella sp. OT286 growing on blood agar plates. This system-level approach would open the possibility of manipulating microbial communities in a targeted fashion as well as associating certain bacterial modules to clinical traits (e.g.: obesity, Crohn's disease, periodontal disease, etc).
Conflict of interest statement
Figures


References
-
- Chung HC, Lee OO, Huang Y-L, Mok SY, Kolter R, et al. Bacterial community succession and chemical profiles of subtidal biofilms in relation to larval settlement of the polychaete Hydroides elegans. ISME J. 2010;4:817–828. - PubMed
-
- Colombo APV, Boches SK, Cotton SL, Goodson JM, Kent R, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. The Journal of Periodontology. 2009;80:1421–1432. - PMC - PubMed
-
- He Z, Xu M, Deng Y, Kang S, Kellogg L, et al. Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecology Letters. 2010;13:564–575. - PubMed
-
- Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Microbial composition of supra- and subgingival plaque in subjects with adult periodontitis. J Clin Periodontol. 2000;27:722–732. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

