Whole genome sequence analysis of Cryptococcus gattii from the Pacific Northwest reveals unexpected diversity
- PMID: 22163313
- PMCID: PMC3233577
- DOI: 10.1371/journal.pone.0028550
Whole genome sequence analysis of Cryptococcus gattii from the Pacific Northwest reveals unexpected diversity
Abstract
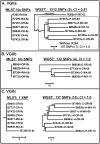
A recent emergence of Cryptococcus gattii in the Pacific Northwest involves strains that fall into three primarily clonal molecular subtypes: VGIIa, VGIIb and VGIIc. Multilocus sequence typing (MLST) and variable number tandem repeat analysis appear to identify little diversity within these molecular subtypes. Given the apparent expansion of these subtypes into new geographic areas and their ability to cause disease in immunocompetent individuals, differentiation of isolates belonging to these subtypes could be very important from a public health perspective. We used whole genome sequence typing (WGST) to perform fine-scale phylogenetic analysis on 20 C. gattii isolates, 18 of which are from the VGII molecular type largely responsible for the Pacific Northwest emergence. Analysis both including and excluding (289,586 SNPs and 56,845 SNPs, respectively) molecular types VGI and VGIII isolates resulted in phylogenetic reconstructions consistent, for the most part, with MLST analysis but with far greater resolution among isolates. The WGST analysis presented here resulted in identification of over 100 SNPs among eight VGIIc isolates as well as unique genotypes for each of the VGIIa, VGIIb and VGIIc isolates. Similar levels of genetic diversity were found within each of the molecular subtype isolates, despite the fact that the VGIIb clade is thought to have emerged much earlier. The analysis presented here is the first multi-genome WGST study to focus on the C. gattii molecular subtypes involved in the Pacific Northwest emergence and describes the tools that will further our understanding of this emerging pathogen.
Conflict of interest statement
Figures


Similar articles
-
Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution.mBio. 2014 Jul 29;5(4):e01494-14. doi: 10.1128/mBio.01494-14. mBio. 2014. PMID: 25073643 Free PMC article.
-
Copy number variation contributes to cryptic genetic variation in outbreak lineages of Cryptococcus gattii from the North American Pacific Northwest.BMC Genomics. 2016 Sep 2;17(1):700. doi: 10.1186/s12864-016-3044-0. BMC Genomics. 2016. PMID: 27590805 Free PMC article.
-
MLST and Whole-Genome-Based Population Analysis of Cryptococcus gattii VGIII Links Clinical, Veterinary and Environmental Strains, and Reveals Divergent Serotype Specific Sub-populations and Distant Ancestors.PLoS Negl Trop Dis. 2016 Aug 5;10(8):e0004861. doi: 10.1371/journal.pntd.0004861. eCollection 2016 Aug. PLoS Negl Trop Dis. 2016. PMID: 27494185 Free PMC article.
-
Cryptococcus gattii comparative genomics and transcriptomics: a NIH/NIAID White Paper.Mycopathologia. 2012 Jun;173(5-6):367-73. doi: 10.1007/s11046-011-9512-9. Epub 2011 Dec 17. Mycopathologia. 2012. PMID: 22179781 Review.
-
Cryptococcus gattii infections.Clin Microbiol Rev. 2014 Oct;27(4):980-1024. doi: 10.1128/CMR.00126-13. Clin Microbiol Rev. 2014. PMID: 25278580 Free PMC article. Review.
Cited by
-
Binding of the wheat germ lectin to Cryptococcus neoformans chitooligomers affects multiple mechanisms required for fungal pathogenesis.Fungal Genet Biol. 2013 Nov;60:64-73. doi: 10.1016/j.fgb.2013.04.005. Epub 2013 Apr 19. Fungal Genet Biol. 2013. PMID: 23608320 Free PMC article.
-
Cryptococcus neoformans Genotypic Diversity and Disease Outcome among HIV Patients in Africa.J Fungi (Basel). 2022 Jul 15;8(7):734. doi: 10.3390/jof8070734. J Fungi (Basel). 2022. PMID: 35887489 Free PMC article. Review.
-
Molecular epidemiology and antifungal susceptibility profiles of clinical Cryptococcus neoformans/Cryptococcus gattii species complex.J Med Microbiol. 2020 Jan;69(1):72-81. doi: 10.1099/jmm.0.001101. J Med Microbiol. 2020. PMID: 31750814 Free PMC article.
-
Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution.mBio. 2014 Jul 29;5(4):e01494-14. doi: 10.1128/mBio.01494-14. mBio. 2014. PMID: 25073643 Free PMC article.
-
Evolutionary role of interspecies hybridization and genetic exchanges in yeasts.Microbiol Mol Biol Rev. 2012 Dec;76(4):721-39. doi: 10.1128/MMBR.00022-12. Microbiol Mol Biol Rev. 2012. PMID: 23204364 Free PMC article. Review.
References
-
- Hoang LM, Maguire JA, Doyle P, Fyfe M, Roscoe DL. Cryptococcus neoformans infections at Vancouver Hospital and Health Sciences Centre (1997–2002): epidemiology, microbiology and histopathology. J Med Microbiol. 2004;53:935–940. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

