Detection of single-nucleotide polymorphism on uidA gene of Escherichia coli by a multiplexed electrochemical DNA biosensor with oligonucleotide-incorporated nonfouling surface
- PMID: 22164059
- PMCID: PMC3231733
- DOI: 10.3390/s110808018
Detection of single-nucleotide polymorphism on uidA gene of Escherichia coli by a multiplexed electrochemical DNA biosensor with oligonucleotide-incorporated nonfouling surface
Abstract
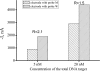
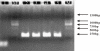
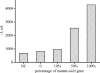
We report here a practical application of a multiplexed electrochemical DNA sensor for highly specific single-nucleotide polymorphism (SNP) detection. In this work, a 16-electrode array was applied with an oligonucleotide-incorporated nonfouling surfaces (ONS) on each electrode for the resistance of unspecific absorption. The fully matched target DNA templated the ligation between the capture probe assembled on gold electrodes and the tandem signal probe with a biotin moiety, which could be transduced to peroxidase-based catalyzed amperometric signals. A mutant site (T93G) in uidA gene of E. coli was analyzed in PCR amplicons. 10% percentage of single mismatched mutant gene was detected, which clearly proved the selectivity of the multiplexed electrochemical DNA biosensor when practically applied.
Keywords: Escherichia coli; electrochemical biosensor; nonfouling electrode surface; single-nucleotide polymorphism (SNP).
Figures
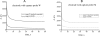
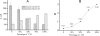
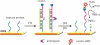
Similar articles
-
Multiplexed electrochemical DNA sensor for single-nucleotide polymorphism typing by using oligonucleotide-incorporated nonfouling surfaces.J Phys Chem B. 2010 May 20;114(19):6703-6. doi: 10.1021/jp100871u. J Phys Chem B. 2010. PMID: 20415492
-
Single-nucleotide polymorphism genotyping using a novel multiplexed electrochemical biosensor with nonfouling surface.Biosens Bioelectron. 2013 Apr 15;42:516-21. doi: 10.1016/j.bios.2012.11.005. Epub 2012 Nov 16. Biosens Bioelectron. 2013. PMID: 23247054
-
Design of an oligonucleotide-incorporated nonfouling surface and its application in electrochemical DNA sensors for highly sensitive and sequence-specific detection of target DNA.Anal Chem. 2008 Dec 1;80(23):9029-33. doi: 10.1021/ac801424y. Anal Chem. 2008. PMID: 19551931
-
Ligase-based multiple DNA analysis by using an electrochemical sensor array.Biosens Bioelectron. 2009 Jan 1;24(5):1209-12. doi: 10.1016/j.bios.2008.07.004. Epub 2008 Jul 12. Biosens Bioelectron. 2009. PMID: 18701273
-
A DNA electrochemical biosensor based on triplex DNA-templated Ag/Pt nanoclusters for the detection of single-nucleotide variant.Talanta. 2020 Jan 15;207:120257. doi: 10.1016/j.talanta.2019.120257. Epub 2019 Aug 14. Talanta. 2020. PMID: 31594620
Cited by
-
Diverse applications of electronic-nose technologies in agriculture and forestry.Sensors (Basel). 2013 Feb 8;13(2):2295-348. doi: 10.3390/s130202295. Sensors (Basel). 2013. PMID: 23396191 Free PMC article. Review.
-
Advances in ligase chain reaction and ligation-based amplifications for genotyping assays: Detection and applications.Mutat Res Rev Mutat Res. 2017 Jul;773:66-90. doi: 10.1016/j.mrrev.2017.05.001. Epub 2017 May 2. Mutat Res Rev Mutat Res. 2017. PMID: 28927538 Free PMC article. Review.
-
Electrochemical Biosensors for Detection of Foodborne Pathogens.Micromachines (Basel). 2019 Mar 28;10(4):222. doi: 10.3390/mi10040222. Micromachines (Basel). 2019. PMID: 30925806 Free PMC article. Review.
References
-
- He Y, Zeng K, Gurung AS, Baloda M, Xu H, Zhang X, Liu G. Visual detection of single-nucleotide polymorphism with hairpin oligonucleotide-functionalized gold nanoparticles. Anal. Chem. 2010;82:7169–7177. - PubMed
-
- Shabo A. Integrating genomics into clinical practice: Standards and regulatory challenges. Curr. Opin. Mol. Ther. 2008;10:267–272. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

