Modes of gene duplication contribute differently to genetic novelty and redundancy, but show parallels across divergent angiosperms
- PMID: 22164235
- PMCID: PMC3229532
- DOI: 10.1371/journal.pone.0028150
Modes of gene duplication contribute differently to genetic novelty and redundancy, but show parallels across divergent angiosperms
Abstract
Background: Both single gene and whole genome duplications (WGD) have recurred in angiosperm evolution. However, the evolutionary effects of different modes of gene duplication, especially regarding their contributions to genetic novelty or redundancy, have been inadequately explored.
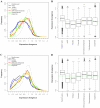
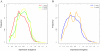
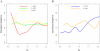
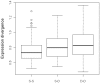
Results: In Arabidopsis thaliana and Oryza sativa (rice), species that deeply sample botanical diversity and for which expression data are available from a wide range of tissues and physiological conditions, we have compared expression divergence between genes duplicated by six different mechanisms (WGD, tandem, proximal, DNA based transposed, retrotransposed and dispersed), and between positional orthologs. Both neo-functionalization and genetic redundancy appear to contribute to retention of duplicate genes. Genes resulting from WGD and tandem duplications diverge slowest in both coding sequences and gene expression, and contribute most to genetic redundancy, while other duplication modes contribute more to evolutionary novelty. WGD duplicates may more frequently be retained due to dosage amplification, while inferred transposon mediated gene duplications tend to reduce gene expression levels. The extent of expression divergence between duplicates is discernibly related to duplication modes, different WGD events, amino acid divergence, and putatively neutral divergence (time), but the contribution of each factor is heterogeneous among duplication modes. Gene loss may retard inter-species expression divergence. Members of different gene families may have non-random patterns of origin that are similar in Arabidopsis and rice, suggesting the action of pan-taxon principles of molecular evolution.
Conclusion: Gene duplication modes differ in contribution to genetic novelty and redundancy, but show some parallels in taxa separated by hundreds of millions of years of evolution.
Conflict of interest statement
Figures




Similar articles
-
Different patterns of gene structure divergence following gene duplication in Arabidopsis.BMC Genomics. 2013 Sep 24;14:652. doi: 10.1186/1471-2164-14-652. BMC Genomics. 2013. PMID: 24063813 Free PMC article.
-
Gene body methylation shows distinct patterns associated with different gene origins and duplication modes and has a heterogeneous relationship with gene expression in Oryza sativa (rice).New Phytol. 2013 Apr;198(1):274-283. doi: 10.1111/nph.12137. Epub 2013 Jan 29. New Phytol. 2013. PMID: 23356482
-
The Evolution of Gene Duplicates in Angiosperms and the Impact of Protein-Protein Interactions and the Mechanism of Duplication.Genome Biol Evol. 2019 Aug 1;11(8):2292-2305. doi: 10.1093/gbe/evz156. Genome Biol Evol. 2019. PMID: 31364708 Free PMC article.
-
Gene duplication as a major force in evolution.J Genet. 2013 Apr;92(1):155-61. doi: 10.1007/s12041-013-0212-8. J Genet. 2013. PMID: 23640422 Review.
-
Consequences of whole genome duplication for 2n pollen performance.Plant Reprod. 2021 Dec;34(4):321-334. doi: 10.1007/s00497-021-00426-z. Epub 2021 Jul 24. Plant Reprod. 2021. PMID: 34302535 Review.
Cited by
-
Comprehensive Genome-Wide Identification of the RNA-Binding Glycine-Rich Gene Family and Expression Profiling under Abiotic Stress in Brassica oleracea.Plants (Basel). 2023 Oct 27;12(21):3706. doi: 10.3390/plants12213706. Plants (Basel). 2023. PMID: 37960062 Free PMC article.
-
Co-expression network analysis of duplicate genes in maize (Zea mays L.) reveals no subgenome bias.BMC Genomics. 2016 Nov 4;17(1):875. doi: 10.1186/s12864-016-3194-0. BMC Genomics. 2016. PMID: 27814670 Free PMC article.
-
Bioinformatics Analysis of the Lipoxygenase Gene Family in Radish (Raphanus sativus) and Functional Characterization in Response to Abiotic and Biotic Stresses.Int J Mol Sci. 2019 Dec 3;20(23):6095. doi: 10.3390/ijms20236095. Int J Mol Sci. 2019. PMID: 31816887 Free PMC article.
-
Epitope-tagged protein-based artificial miRNA screens for optimized gene silencing in plants.Nat Protoc. 2014 Apr;9(4):939-49. doi: 10.1038/nprot.2014.061. Epub 2014 Mar 27. Nat Protoc. 2014. PMID: 24675734 Free PMC article.
-
Recent emergence and extinction of the protection of telomeres 1c gene in Arabidopsis thaliana.Plant Cell Rep. 2019 Sep;38(9):1081-1097. doi: 10.1007/s00299-019-02427-9. Epub 2019 May 27. Plant Cell Rep. 2019. PMID: 31134349 Free PMC article.
References
-
- Paterson AH, Freeling M, Tang H, Wang X. Insights from the comparison of plant genome sequences. Annu Rev Plant Biol. 2010;61:349–372. - PubMed
-
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. - PubMed
-
- Aury JM, Jaillon O, Duret L, Noel B, Jubin C, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. - PubMed
-
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. - PubMed
-
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

