Biochemically defined HIV-1 envelope glycoprotein variant immunogens display differential binding and neutralizing specificities to the CD4-binding site
- PMID: 22167180
- PMCID: PMC3285340
- DOI: 10.1074/jbc.M111.317776
Biochemically defined HIV-1 envelope glycoprotein variant immunogens display differential binding and neutralizing specificities to the CD4-binding site
Abstract
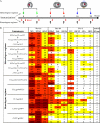
HIV-1 gp120 binds the primary receptor CD4. Recently, a plethora of broadly neutralizing antibodies to the gp120 CD4-binding site (CD4bs) validated this region as a target for immunogen design. Here, we asked if modified HIV-1 envelope glycoproteins (Env) designed to increase CD4 recognition might improve recognition by CD4bs neutralizing antibodies and more efficiently elicit such reactivities. We also asked if CD4bs stabilization, coupled with altering the Env format (monomer to trimer or cross-clade), might better elicit neutralizing antibodies by focusing the immune response on the functionally conserved CD4bs. We produced monomeric and trimeric Envs stabilized by mutations within the gp120 CD4bs cavity (pocket-filling; PF2) or by appending heterologous trimerization motifs to soluble Env ectodomains (gp120/gp140). Recombinant glycoproteins were purified to relative homogeneity, and ligand binding properties were analyzed by ELISA, surface plasmon resonance, and isothermal titration microcalorimetry. In some formats, the PF2 substitutions increased CD4 affinity, and importantly, PF2-containing proteins were better recognized by the broadly neutralizing CD4bs mAbs, VRC01 and VRC-PG04. Based on this analysis, we immunized selected Env variants into rabbits using heterologous or homologous regimens. Analysis of the sera revealed that homologous inoculation of the PF2-containing, variable region-deleted YU2 gp120 trimers (ΔV123/PF2-GCN4) more rapidly elicited CD4bs-directed neutralizing antibodies compared with other regimens, whereas homologous trimers elicited increased neutralization potency, mapping predominantly to the gp120 third major variable region (V3). These results suggest that some engineered Env proteins may more efficiently direct responses toward the conserved CD4bs and be valuable to elicit antibodies of greater neutralizing capacity.
Figures


Similar articles
-
Hyperglycosylated stable core immunogens designed to present the CD4 binding site are preferentially recognized by broadly neutralizing antibodies.J Virol. 2014 Dec;88(24):14002-16. doi: 10.1128/JVI.02614-14. Epub 2014 Sep 24. J Virol. 2014. PMID: 25253346 Free PMC article.
-
PGV04, an HIV-1 gp120 CD4 binding site antibody, is broad and potent in neutralization but does not induce conformational changes characteristic of CD4.J Virol. 2012 Apr;86(8):4394-403. doi: 10.1128/JVI.06973-11. Epub 2012 Feb 15. J Virol. 2012. PMID: 22345481 Free PMC article.
-
An HIV-1 Env-Antibody Complex Focuses Antibody Responses to Conserved Neutralizing Epitopes.J Immunol. 2016 Nov 15;197(10):3982-3998. doi: 10.4049/jimmunol.1601134. Epub 2016 Oct 10. J Immunol. 2016. PMID: 27815444 Free PMC article.
-
Quaternary Interaction of the HIV-1 Envelope Trimer with CD4 and Neutralizing Antibodies.Viruses. 2021 Jul 20;13(7):1405. doi: 10.3390/v13071405. Viruses. 2021. PMID: 34372611 Free PMC article. Review.
-
Elicitation of HIV-1-neutralizing antibodies against the CD4-binding site.Curr Opin HIV AIDS. 2013 Sep;8(5):382-92. doi: 10.1097/COH.0b013e328363a90e. Curr Opin HIV AIDS. 2013. PMID: 23924998 Free PMC article. Review.
Cited by
-
Design of an Escherichia coli expressed HIV-1 gp120 fragment immunogen that binds to b12 and induces broad and potent neutralizing antibodies.J Biol Chem. 2013 Apr 5;288(14):9815-9825. doi: 10.1074/jbc.M112.425959. Epub 2013 Feb 21. J Biol Chem. 2013. PMID: 23430741 Free PMC article.
-
Diverse antibody genetic and recognition properties revealed following HIV-1 envelope glycoprotein immunization.J Immunol. 2015 Jun 15;194(12):5903-14. doi: 10.4049/jimmunol.1500122. Epub 2015 May 11. J Immunol. 2015. PMID: 25964491 Free PMC article.
-
Increased functional stability and homogeneity of viral envelope spikes through directed evolution.PLoS Pathog. 2013 Feb;9(2):e1003184. doi: 10.1371/journal.ppat.1003184. Epub 2013 Feb 28. PLoS Pathog. 2013. PMID: 23468626 Free PMC article.
-
Pre-clinical development of a recombinant, replication-competent adenovirus serotype 4 vector vaccine expressing HIV-1 envelope 1086 clade C.PLoS One. 2013 Dec 3;8(12):e82380. doi: 10.1371/journal.pone.0082380. eCollection 2013. PLoS One. 2013. PMID: 24312658 Free PMC article.
-
HIV-1 clade C escapes broadly neutralizing autologous antibodies with N332 glycan specificity by distinct mechanisms.Retrovirology. 2016 Aug 30;13(1):60. doi: 10.1186/s12977-016-0297-2. Retrovirology. 2016. PMID: 27576440 Free PMC article.
References
-
- Lu M., Blacklow S. C., Kim P. S. (1995) A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2, 1075–1082 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

