REF4 and RFR1, subunits of the transcriptional coregulatory complex mediator, are required for phenylpropanoid homeostasis in Arabidopsis
- PMID: 22167189
- PMCID: PMC3285322
- DOI: 10.1074/jbc.M111.312298
REF4 and RFR1, subunits of the transcriptional coregulatory complex mediator, are required for phenylpropanoid homeostasis in Arabidopsis
Abstract
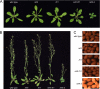
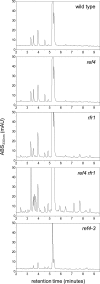
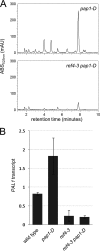
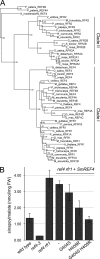
The plant phenylpropanoid pathway produces an array of metabolites that impact human health and the utility of feed and fiber crops. We previously characterized several Arabidopsis thaliana mutants with dominant mutations in REDUCED EPIDERMAL FLUORESCENCE 4 (REF4) that cause dwarfing and decreased accumulation of phenylpropanoids. In contrast, ref4 null plants are of normal stature and have no apparent defect in phenylpropanoid biosynthesis. Here we show that disruption of both REF4 and its paralog, REF4-RELATED 1 (RFR1), results in enhanced expression of multiple phenylpropanoid biosynthetic genes, as well as increased accumulation of numerous downstream products. We also show that the dominant ref4-3 mutant protein interferes with the ability of the PAP1/MYB75 transcription factor to induce the expression of PAL1 and drive anthocyanin accumulation. Consistent with our experimental results, both REF4 and RFR1 have been shown to physically associate with the conserved transcriptional coregulatory complex, Mediator, which transduces information from cis-acting DNA elements to RNA polymerase II at the core promoter. Taken together, our data provide critical genetic support for a functional role of REF4 and RFR1 in the Mediator complex, and for Mediator in the maintenance of phenylpropanoid homeostasis. Finally, we show that wild-type RFR1 substantially mitigates the phenotype of the dominant ref4-3 mutant, suggesting that REF4 and RFR1 may compete with one another for common binding partners or for occupancy in Mediator. Determining the functions of diverse Mediator subunits is essential to understand eukaryotic gene regulation, and to facilitate rational manipulation of plant metabolic pathways to better suit human needs.
Figures
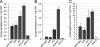
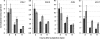

Similar articles
-
The function of the Mediator complex in plant immunity.Plant Signal Behav. 2013 Mar;8(3):e23182. doi: 10.4161/psb.23182. Epub 2013 Jan 8. Plant Signal Behav. 2013. PMID: 23299323 Free PMC article. Review.
-
Mediator Complex Subunits MED2, MED5, MED16, and MED23 Genetically Interact in the Regulation of Phenylpropanoid Biosynthesis.Plant Cell. 2017 Dec;29(12):3269-3285. doi: 10.1105/tpc.17.00282. Epub 2017 Dec 4. Plant Cell. 2017. PMID: 29203634 Free PMC article.
-
Semidominant mutations in reduced epidermal fluorescence 4 reduce phenylpropanoid content in Arabidopsis.Genetics. 2008 Apr;178(4):2237-51. doi: 10.1534/genetics.107.083881. Genetics. 2008. PMID: 18430946 Free PMC article.
-
Mutation of Mediator subunit CDK8 counteracts the stunted growth and salicylic acid hyperaccumulation phenotypes of an Arabidopsis MED5 mutant.New Phytol. 2019 Jul;223(1):233-245. doi: 10.1111/nph.15741. Epub 2019 Mar 18. New Phytol. 2019. PMID: 30756399
-
A semidominant point mutation of Mediator tail subunit MED5b in Arabidopsis leads to altered enrichment of H3K27me3 and reduced expression of targets of MYC2.G3 (Bethesda). 2025 Mar 18;15(3):jkae301. doi: 10.1093/g3journal/jkae301. G3 (Bethesda). 2025. PMID: 39950577 Free PMC article.
Cited by
-
The function of the Mediator complex in plant immunity.Plant Signal Behav. 2013 Mar;8(3):e23182. doi: 10.4161/psb.23182. Epub 2013 Jan 8. Plant Signal Behav. 2013. PMID: 23299323 Free PMC article. Review.
-
Transcriptome Analysis of Four Arabidopsis thaliana Mediator Tail Mutants Reveals Overlapping and Unique Functions in Gene Regulation.G3 (Bethesda). 2018 Aug 30;8(9):3093-3108. doi: 10.1534/g3.118.200573. G3 (Bethesda). 2018. PMID: 30049745 Free PMC article.
-
Towards a Cardoon (Cynara cardunculus var. altilis)-Based Biorefinery: A Case Study of Improved Cell Cultures via Genetic Modulation of the Phenylpropanoid Pathway.Int J Mol Sci. 2021 Nov 5;22(21):11978. doi: 10.3390/ijms222111978. Int J Mol Sci. 2021. PMID: 34769407 Free PMC article.
-
Mapping the Gene Expression Spectrum of Mediator Subunits in Response to Viroid Infection in Plants.Int J Mol Sci. 2020 Apr 3;21(7):2498. doi: 10.3390/ijms21072498. Int J Mol Sci. 2020. PMID: 32260277 Free PMC article.
-
Transcriptional responses of wheat roots inoculated with Arthrobacter nitroguajacolicus to salt stress.Sci Rep. 2019 Feb 11;9(1):1792. doi: 10.1038/s41598-018-38398-2. Sci Rep. 2019. PMID: 30741989 Free PMC article.
References
-
- Bonawitz N. D., Chapple C. (2010) The genetics of lignin biosynthesis. Connecting genotype to phenotype. Annu. Rev. Genet. 44, 337–363 - PubMed
-
- Ralph J. (2004) Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem. Rev. 3, 29–60
-
- Field C. B., Behrenfeld M. J., Randerson J. T., Falkowski P. (1998) Primary production of the biosphere. Integrating terrestrial and oceanic components. Science 281, 237–240 - PubMed
-
- Umezawa T. (2010) The cinnamate/monolignol pathway. Phytochem. Rev. 9, 1–17
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

