Identification of the heparin binding site on adeno-associated virus serotype 3B (AAV-3B)
- PMID: 22169623
- PMCID: PMC3253896
- DOI: 10.1016/j.virol.2011.10.007
Identification of the heparin binding site on adeno-associated virus serotype 3B (AAV-3B)
Abstract
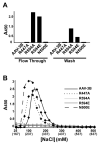
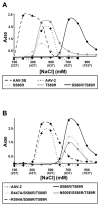
Adeno-associated virus is a promising vector for gene therapy. In the current study, the binding site on AAV serotype 3B for the heparan sulfate proteoglycan (HSPG) receptor has been characterized. X-ray diffraction identified a disaccharide binding site at the most positively charged region on the virus surface. The contributions of basic amino acids at this and other sites were characterized using site-directed mutagenesis. Both heparin and cell binding are correlated to positive charge at the disaccharide binding site, and transduction is significantly decreased in AAV-3B vectors mutated at this site to reduce heparin binding. While the receptor attachment sites of AAV-3B and AAV-2 are both in the general vicinity of the viral spikes, the exact amino acids that participate in electrostatic interactions are distinct. Diversity in the mechanisms of cell attachment by AAV serotypes will be an important consideration for the rational design of improved gene therapy vectors.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Structure-function analysis of receptor-binding in adeno-associated virus serotype 6 (AAV-6).Virology. 2011 Nov 10;420(1):10-9. doi: 10.1016/j.virol.2011.08.011. Epub 2011 Sep 13. Virology. 2011. PMID: 21917284 Free PMC article.
-
Heparan sulfate proteoglycan affinity of adeno-associated virus vectors: Implications for retinal gene delivery.Eur J Pharm Sci. 2025 Mar 1;206:107012. doi: 10.1016/j.ejps.2025.107012. Epub 2025 Jan 11. Eur J Pharm Sci. 2025. PMID: 39805508
-
Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding.J Virol. 2003 Jun;77(12):6995-7006. doi: 10.1128/jvi.77.12.6995-7006.2003. J Virol. 2003. PMID: 12768018 Free PMC article.
-
Structural and cellular biology of adeno-associated virus attachment and entry.Adv Virus Res. 2020;106:39-84. doi: 10.1016/bs.aivir.2020.01.002. Epub 2020 Feb 13. Adv Virus Res. 2020. PMID: 32327148 Review.
-
Adeno-Associated Virus (AAV) Gene Delivery: Dissecting Molecular Interactions upon Cell Entry.Viruses. 2021 Jul 10;13(7):1336. doi: 10.3390/v13071336. Viruses. 2021. PMID: 34372542 Free PMC article. Review.
Cited by
-
Adeno-associated viral vectors based on serotype 3b use components of the fibroblast growth factor receptor signaling complex for efficient transduction.Hum Gene Ther. 2012 Oct;23(10):1031-42. doi: 10.1089/hum.2012.066. Epub 2012 Aug 27. Hum Gene Ther. 2012. PMID: 22680698 Free PMC article.
-
AAV13 Enables Precise Targeting of Local Neural Populations.Int J Mol Sci. 2022 Oct 24;23(21):12806. doi: 10.3390/ijms232112806. Int J Mol Sci. 2022. PMID: 36361595 Free PMC article.
-
Hexon modification of human adenovirus type 5 vectors enables efficient transduction of human multipotent mesenchymal stromal cells.Mol Ther Methods Clin Dev. 2022 Mar 7;25:96-110. doi: 10.1016/j.omtm.2022.03.004. eCollection 2022 Jun 9. Mol Ther Methods Clin Dev. 2022. PMID: 35402633 Free PMC article.
-
Parvovirus Capsid-Antibody Complex Structures Reveal Conservation of Antigenic Epitopes Across the Family.Viral Immunol. 2021 Jan-Feb;34(1):3-17. doi: 10.1089/vim.2020.0022. Epub 2020 Apr 21. Viral Immunol. 2021. PMID: 32315582 Free PMC article. Review.
-
Structure of AAV-DJ, a retargeted gene therapy vector: cryo-electron microscopy at 4.5 Å resolution.Structure. 2012 Aug 8;20(8):1310-20. doi: 10.1016/j.str.2012.05.004. Epub 2012 Jun 21. Structure. 2012. PMID: 22727812 Free PMC article.
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–54. - PubMed
-
- Asokan A, Conway JC, Phillips JL, Li C, Hegge J, Sinnott R, Yadav S, DiPrimio N, Nam HJ, Agbandje-McKenna M, McPhee S, Wolff J, Samulski RJ. Reengineering a receptor footprint of adeno-associated virus enables selective and systemic gene transfer to muscle. Nat Biotechnol. 2010;28(1):79–82. - PMC - PubMed
-
- Ayuso E, Mingozzi F, Montane J, Leon X, Anguela XM, Haurigot V, Edmonson SA, Africa L, Zhou S, High KA, Bosch F, Wright JF. High AAV vector purity results in serotype- and tissue-independent enhancement of transduction efficiency. Gene Ther. 17(4):503–10. - PubMed
-
- Badger J, Minor I, Kremer M, Oliveira M, Smith TJ, Griffith JP, Guerin DM, Krishnaswarmy S, Luo M, Rossmann MG, McKinlay M, Diana G, Dutko FJ, Fancher M, Rueckert R, Heinz BA. Structural Analysis of a series of antiviral agents complexed with human rhinovirus 14. Proceedings of the National Academy of Sciences, USA. 1988;85:3304–3308. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

