Subunit interactions and organization of the Chlamydomonas reinhardtii intraflagellar transport complex A proteins
- PMID: 22170070
- PMCID: PMC3320918
- DOI: 10.1074/jbc.M111.287102
Subunit interactions and organization of the Chlamydomonas reinhardtii intraflagellar transport complex A proteins
Abstract
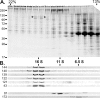
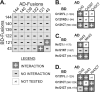
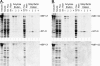
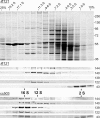
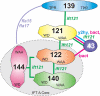
Chlamydomonas reinhardtii intraflagellar transport (IFT) particles can be biochemically resolved into two smaller assemblies, complexes A and B, that contain up to six and 15 protein subunits, respectively. We provide here the proteomic and immunological analyses that verify the identity of all six Chlamydomonas A proteins. Using sucrose density gradient centrifugation and antibody pulldowns, we show that all six A subunits are associated in a 16 S complex in both the cell bodies and flagella. A significant fraction of the cell body IFT43, however, exhibits a much slower sedimentation of ∼2 S and is not associated with the IFT A complex. To identify interactions between the six A proteins, we combined exhaustive yeast-based two-hybrid analysis, heterologous recombinant protein expression in Escherichia coli, and analysis of the newly identified complex A mutants, ift121 and ift122. We show that IFT121 and IFT43 interact directly and provide evidence for additional interactions between IFT121 and IFT139, IFT121 and IFT122, IFT140 and IFT122, and IFT140 and IFT144. The mutant analysis further allows us to propose that a subset of complex A proteins, IFT144/140/122, can form a stable 12 S subcomplex that we refer to as the IFT A core. Based on these results, we propose a model for the spatial arrangement of the six IFT A components.
Figures



References
-
- Afzelius B. A. (1995) Role of cilia in human health. Cell Motil. Cytoskeleton. 32, 95–97 - PubMed
-
- Vincensini L., Blisnick T., Bastin P. (2011) 1001 model organisms to study cilia and flagella. Biol. Cell 103, 109–130 - PubMed
-
- Corbit K. C., Aanstad P., Singla V., Norman A. R., Stainier D. Y., Reiter J. F. (2005) Vertebrate Smoothened functions at the primary cilium. Nature 437, 1018–1021 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

