A metagenome of a full-scale microbial community carrying out enhanced biological phosphorus removal
- PMID: 22170425
- PMCID: PMC3358022
- DOI: 10.1038/ismej.2011.176
A metagenome of a full-scale microbial community carrying out enhanced biological phosphorus removal
Abstract
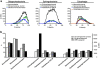
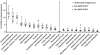
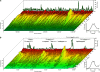
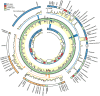
Enhanced biological phosphorus removal (EBPR) is widely used for removal of phosphorus from wastewater. In this study, a metagenome (18.2 Gb) was generated using Illumina sequencing from a full-scale EBPR plant to study the community structure and genetic potential. Quantitative fluorescence in situ hybridization (qFISH) was applied as an independent method to evaluate the community structure. The results were in qualitative agreement, but a DNA extraction bias against gram positive bacteria using standard extraction protocols was identified, which would not have been identified without the use of qFISH. The genetic potential for community function showed enrichment of genes involved in phosphate metabolism and biofilm formation, reflecting the selective pressure of the EBPR process. Most contigs in the assembled metagenome had low similarity to genes from currently sequenced genomes, underlining the need for more reference genomes of key EBPR species. Only the genome of 'Candidatus Accumulibacter', a genus of phosphorus-removing organisms, was closely enough related to the species present in the metagenome to allow for detailed investigations. Accumulibacter accounted for only 4.8% of all bacteria by qFISH, but the depth of sequencing enabled detailed insight into their microdiversity in the full-scale plant. Only 15% of the reads matching Accumulibacter had a high similarity (>95%) to the sequenced Accumulibacter clade IIA strain UW-1 genome, indicating the presence of some microdiversity. The differences in gene complement between the Accumulibacter clades were limited to genes for extracellular polymeric substances and phage-related genes, suggesting a selective pressure from phages on the Accumulibacter diversity.
Figures

References
-
- Barr JJ, Slater FR, Fukushima T, Bond PL. Evidence for bacteriophage activity causing community and performance changes in a phosphorus-removal activated sludge. FEMS Microbiol Ecol. 2010;74:631–642. - PubMed
-
- Brazelton WJ, Baross JA. Abundant transposases encoded by the metagenome of a hydrothermal chimney biofilm. ISME J. 2009;3:1420–1424. - PubMed

