Development and validation of a comparative genomic fingerprinting method for high-resolution genotyping of Campylobacter jejuni
- PMID: 22170908
- PMCID: PMC3295178
- DOI: 10.1128/JCM.00669-11
Development and validation of a comparative genomic fingerprinting method for high-resolution genotyping of Campylobacter jejuni
Abstract
Campylobacter spp. are a leading cause of bacterial gastroenteritis worldwide. The need for molecular subtyping methods with enhanced discrimination in the context of surveillance- and outbreak-based epidemiologic investigations of Campylobacter spp. is critical to our understanding of sources and routes of transmission and the development of mitigation strategies to reduce the incidence of campylobacteriosis. We describe the development and validation of a rapid and high-resolution comparative genomic fingerprinting (CGF) method for C. jejuni. A total of 412 isolates from agricultural, environmental, retail, and human clinical sources obtained from the Canadian national integrated enteric pathogen surveillance program (C-EnterNet) were analyzed using a 40-gene assay (CGF40) and multilocus sequence typing (MLST). The significantly higher Simpson's index of diversity (ID) obtained with CGF40 (ID = 0.994) suggests that it has a higher discriminatory power than MLST at both the level of clonal complex (ID = 0.873) and sequence type (ID = 0.935). High Wallace coefficients obtained when CGF40 was used as the primary typing method suggest that CGF and MLST are highly concordant, and we show that isolates with identical MLST profiles are comprised of isolates with distinct but highly similar CGF profiles. The high concordance with MLST coupled with the ability to discriminate between closely related isolates suggests that CFG40 is useful in differentiating highly prevalent sequence types, such as ST21 and ST45. CGF40 is a high-resolution comparative genomics-based method for C. jejuni subtyping with high discriminatory power that is also rapid, low cost, and easily deployable for routine epidemiologic surveillance and outbreak investigations.
Figures

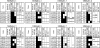
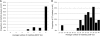
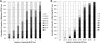
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

