First worldwide proficiency study on variable-number tandem-repeat typing of Mycobacterium tuberculosis complex strains
- PMID: 22170917
- PMCID: PMC3295139
- DOI: 10.1128/JCM.00607-11
First worldwide proficiency study on variable-number tandem-repeat typing of Mycobacterium tuberculosis complex strains
Abstract
Although variable-number tandem-repeat (VNTR) typing has gained recognition as the new standard for the DNA fingerprinting of Mycobacterium tuberculosis complex (MTBC) isolates, external quality control programs have not yet been developed. Therefore, we organized the first multicenter proficiency study on 24-locus VNTR typing. Sets of 30 DNAs of MTBC strains, including 10 duplicate DNA samples, were distributed among 37 participating laboratories in 30 different countries worldwide. Twenty-four laboratories used an in-house-adapted method with fragment sizing by gel electrophoresis or an automated DNA analyzer, nine laboratories used a commercially available kit, and four laboratories used other methods. The intra- and interlaboratory reproducibilities of VNTR typing varied from 0% to 100%, with averages of 72% and 60%, respectively. Twenty of the 37 laboratories failed to amplify particular VNTR loci; if these missing results were ignored, the number of laboratories with 100% interlaboratory reproducibility increased from 1 to 5. The average interlaboratory reproducibility of VNTR typing using a commercial kit was better (88%) than that of in-house-adapted methods using a DNA analyzer (70%) or gel electrophoresis (50%). Eleven laboratories using in-house-adapted manual typing or automated typing scored inter- and intralaboratory reproducibilities of 80% or higher, which suggests that these approaches can be used in a reliable way. In conclusion, this first multicenter study has documented the worldwide quality of VNTR typing of MTBC strains and highlights the importance of international quality control to improve genotyping in the future.
Figures
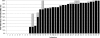
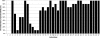

Comment in
-
Evaluation of mycobacterial interspersed repetitive-unit-variable-number tandem-repeat genotyping as performed in laboratories in Canada, France, and the United States.J Clin Microbiol. 2012 May;50(5):1830-1; author reply 1832. doi: 10.1128/JCM.00168-12. J Clin Microbiol. 2012. PMID: 22505766 Free PMC article. No abstract available.
References
-
- Allix C, Supply P, Fauville-Dufaux M. 2004. Utility of fast mycobacterial interspersed repetitive unit-variable number tandem repeat genotyping in clinical mycobacteriological analysis. Clin. Infect. Dis. 39:783–789 - PubMed
-
- Brown TJ, Nikolayevskyy VN, Drobniewski FA. 2009. Typing Mycobacterium tuberculosis using variable number tandem repeat analysis. Methods Mol. Biol. 465:371–394 - PubMed
-
- Cole ST, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

