Photo-ribonucleotide reductase β2 by selective cysteine labeling with a radical phototrigger
- PMID: 22171005
- PMCID: PMC3252915
- DOI: 10.1073/pnas.1115778108
Photo-ribonucleotide reductase β2 by selective cysteine labeling with a radical phototrigger
Abstract
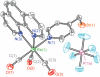
Photochemical radical initiation is a powerful tool for studying radical initiation and transport in biology. Ribonucleotide reductases (RNRs), which catalyze the conversion of nucleotides to deoxynucleotides in all organisms, are an exemplar of radical mediated transformations in biology. Class Ia RNRs are composed of two subunits: α2 and β2. As a method to initiate radical formation photochemically within β2, a single surface-exposed cysteine of the β2 subunit of Escherichia coli Class Ia RNR has been labeled (98%) with a photooxidant ([Re ] = tricarbonyl(1,10-phenanthroline)(methylpyridyl)rhenium(I)). The labeling was achieved by incubation of S355C-β2 with the 4-(bromomethyl)pyridyl derivative of [Re] to yield the labeled species, [Re]-S355C-β2. Steady-state and time-resolved emission experiments reveal that the metal-to-ligand charge transfer (MLCT) excited-state (3)[Re ](∗) is not significantly perturbed after bioconjugation and is available as a phototrigger of tyrosine radical at position 356 in the β2 subunit; transient absorption spectroscopy reveals that the radical lives for microseconds. The work described herein provides a platform for photochemical radical initiation and study of proton-coupled electron transfer (PCET) in the β2 subunit of RNR, from which radical initiation and transport for this enzyme originates.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Reversible, long-range radical transfer in E. coli class Ia ribonucleotide reductase.Acc Chem Res. 2013 Nov 19;46(11):2524-35. doi: 10.1021/ar4000407. Epub 2013 Jun 4. Acc Chem Res. 2013. PMID: 23730940 Free PMC article.
-
A Proton Wire Mediates Proton Coupled Electron Transfer from Hydroxyurea and Other Hydroxamic Acids to Tyrosyl Radical in Class Ia Ribonucleotide Reductase.J Phys Chem B. 2020 Jan 16;124(2):345-354. doi: 10.1021/acs.jpcb.9b08587. Epub 2020 Jan 6. J Phys Chem B. 2020. PMID: 31904962
-
Kinetics of hydrogen atom abstraction from substrate by an active site thiyl radical in ribonucleotide reductase.J Am Chem Soc. 2014 Nov 19;136(46):16210-6. doi: 10.1021/ja507313w. Epub 2014 Nov 10. J Am Chem Soc. 2014. PMID: 25353063 Free PMC article.
-
Long-range proton-coupled electron transfer in the Escherichia coli class Ia ribonucleotide reductase.Essays Biochem. 2017 May 9;61(2):281-292. doi: 10.1042/EBC20160072. Print 2017 May 9. Essays Biochem. 2017. PMID: 28487404 Review.
-
The prototypic class Ia ribonucleotide reductase from Escherichia coli: still surprising after all these years.Biochem Soc Trans. 2012 Jun 1;40(3):523-30. doi: 10.1042/BST20120081. Biochem Soc Trans. 2012. PMID: 22616862 Free PMC article. Review.
Cited by
-
Reversible phenol oxidation and reduction in the structurally well-defined 2-Mercaptophenol-α₃C protein.Biochemistry. 2013 Feb 26;52(8):1409-18. doi: 10.1021/bi301613p. Epub 2013 Feb 14. Biochemistry. 2013. PMID: 23373469 Free PMC article.
-
High-Frequency/High-Field Electron Paramagnetic Resonance and Theoretical Studies of Tryptophan-Based Radicals.J Phys Chem A. 2018 Mar 29;122(12):3170-3176. doi: 10.1021/acs.jpca.7b12434. Epub 2018 Mar 15. J Phys Chem A. 2018. PMID: 29488750 Free PMC article.
-
Reversible, long-range radical transfer in E. coli class Ia ribonucleotide reductase.Acc Chem Res. 2013 Nov 19;46(11):2524-35. doi: 10.1021/ar4000407. Epub 2013 Jun 4. Acc Chem Res. 2013. PMID: 23730940 Free PMC article.
-
Catalysis and Electron Transfer in De Novo Designed Helical Scaffolds.Angew Chem Int Ed Engl. 2020 May 11;59(20):7678-7699. doi: 10.1002/anie.201907502. Epub 2020 Mar 2. Angew Chem Int Ed Engl. 2020. PMID: 31441170 Free PMC article. Review.
-
Kinetic model for reversible radical transfer in ribonucleotide reductase.Proc Natl Acad Sci U S A. 2022 Jun 21;119(25):e2202022119. doi: 10.1073/pnas.2202022119. Epub 2022 Jun 17. Proc Natl Acad Sci U S A. 2022. PMID: 35714287 Free PMC article.
References
-
- Reece SY, Nocera DG. In: Quantum Tunnelling in Enzyme-Catalysed Reactions. Scrutton NS, Allemann RK, editors. London: Royal Society of Chemistry Press; 2009. pp. 345–377.
-
- Hammes-Schiffer S. Hydrogen tunneling and protein motion in enzyme reactions. Acc Chem Res. 2006;39:93–100. - PubMed
-
- Liang Z, Klinman JP. Structural bases of hydrogen tunneling in enzymes: progress and puzzles. Curr Opin Struct Biol. 2004;14:648–655. - PubMed
-
- Van der Donk WA, Stubbe J. Protein radicals in enzyme catalysis. Chem Rev. 1998;98:705–762. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

