Roles of the Drosophila LRRK2 homolog in Rab7-dependent lysosomal positioning
- PMID: 22171073
- PMCID: PMC3284123
- DOI: 10.1093/hmg/ddr573
Roles of the Drosophila LRRK2 homolog in Rab7-dependent lysosomal positioning
Abstract
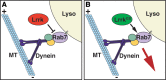
LRRK2 (PARK8) is the most common genetic determinant of Parkinson's disease (PD), with dominant mutations in LRRK2 causing inherited PD and sequence variation at the LRRK2 locus associated with increased risk for sporadic PD. Although LRRK2 has been implicated in diverse cellular processes encompassing almost all cellular compartments, the precise functions of LRRK2 remain unclear. Here, we show that the Drosophila homolog of LRRK2 (Lrrk) localizes to the membranes of late endosomes and lysosomes, physically interacts with the crucial mediator of late endosomal transport Rab7 and negatively regulates rab7-dependent perinuclear localization of lysosomes. We also show that a mutant form of lrrk analogous to the pathogenic LRRK2(G2019S) allele behaves oppositely to wild-type lrrk in that it promotes rather than inhibits rab7-dependent perinuclear lysosome clustering, with these effects of mutant lrrk on lysosome position requiring both microtubules and dynein. These data suggest that LRRK2 normally functions in Rab7-dependent lysosomal positioning, and that this function is disrupted by the most common PD-causing LRRK2 mutation, linking endolysosomal dysfunction to the pathogenesis of LRRK2-mediated PD.
Figures




Similar articles
-
Novel ethyl methanesulfonate (EMS)-induced null alleles of the Drosophila homolog of LRRK2 reveal a crucial role in endolysosomal functions and autophagy in vivo.Dis Model Mech. 2014 Dec;7(12):1351-63. doi: 10.1242/dmm.017020. Epub 2014 Oct 2. Dis Model Mech. 2014. PMID: 25288684 Free PMC article.
-
LRRK2 delays degradative receptor trafficking by impeding late endosomal budding through decreasing Rab7 activity.Hum Mol Genet. 2014 Dec 20;23(25):6779-96. doi: 10.1093/hmg/ddu395. Epub 2014 Jul 30. Hum Mol Genet. 2014. PMID: 25080504
-
The G2019S variant of leucine-rich repeat kinase 2 (LRRK2) alters endolysosomal trafficking by impairing the function of the GTPase RAB8A.J Biol Chem. 2019 Mar 29;294(13):4738-4758. doi: 10.1074/jbc.RA118.005008. Epub 2019 Feb 1. J Biol Chem. 2019. PMID: 30709905 Free PMC article.
-
The synaptic function of LRRK2.Biochem Soc Trans. 2012 Oct;40(5):1047-51. doi: 10.1042/BST20120113. Biochem Soc Trans. 2012. PMID: 22988863 Review.
-
LRRK2 and the Endolysosomal System in Parkinson's Disease.J Parkinsons Dis. 2020;10(4):1271-1291. doi: 10.3233/JPD-202138. J Parkinsons Dis. 2020. PMID: 33044192 Free PMC article. Review.
Cited by
-
Lrrk regulates the dynamic profile of dendritic Golgi outposts through the golgin Lava lamp.J Cell Biol. 2015 Aug 3;210(3):471-83. doi: 10.1083/jcb.201411033. Epub 2015 Jul 27. J Cell Biol. 2015. PMID: 26216903 Free PMC article.
-
Function and dysfunction of leucine-rich repeat kinase 2 (LRRK2): Parkinson's disease and beyond.BMB Rep. 2015 May;48(5):243-8. doi: 10.5483/bmbrep.2015.48.5.032. BMB Rep. 2015. PMID: 25703537 Free PMC article. Review.
-
Mechanisms of LRRK2-mediated neurodegeneration.Curr Neurol Neurosci Rep. 2012 Jun;12(3):251-60. doi: 10.1007/s11910-012-0265-8. Curr Neurol Neurosci Rep. 2012. PMID: 22441981 Review.
-
Autophagy and LRRK2 in the Aging Brain.Front Neurosci. 2019 Dec 17;13:1352. doi: 10.3389/fnins.2019.01352. eCollection 2019. Front Neurosci. 2019. PMID: 31920513 Free PMC article. Review.
-
Novel ethyl methanesulfonate (EMS)-induced null alleles of the Drosophila homolog of LRRK2 reveal a crucial role in endolysosomal functions and autophagy in vivo.Dis Model Mech. 2014 Dec;7(12):1351-63. doi: 10.1242/dmm.017020. Epub 2014 Oct 2. Dis Model Mech. 2014. PMID: 25288684 Free PMC article.
References
-
- Paisan-Ruiz C., Jain S., Evans E.W., Gilks W.P., Simon J., van der Brug M., Lopz de Munain A., Aparicio S., Gil A.M., Khan N., et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi:10.1016/j.neuron.2004.10.023. - DOI - PubMed
-
- Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R.J., Calne D.B., et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi:10.1016/j.neuron.2004.11.005. - DOI - PubMed
-
- Satake W., Nakabayashi Y., Mizuta I., Hirota Y., Ito C., Kubo M., Kawaguchi T., Tsunoda T., Watanabe M., Takeda A., et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat. Genet. 2009;41:1303–1307. doi:10.1038/ng.485. - DOI - PubMed
-
- Simon-Sanchez J., Schulte C., Bras J.M., Sharma M., Gibbs J.R., Berg D., Paisan-Ruiz C., Lichtner P., Scholz S.W., Hernandez D.G., et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat. Genet. 2009;41:1308–1312. doi:10.1038/ng.487. - DOI - PMC - PubMed
-
- Lesage S., Ibanez P., Lohmann E., Pollak P., Tison F., Tazir M., Leutenegger A.L., Guimaraes J., Bonnet A.M., Agid Y., et al. G2019S LRRK2 mutation in French and North African families with Parkinson's disease. Ann. Neurol. 2005;58:784–787. doi:10.1002/ana.20636. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

